Juvenile hormone epoxide hydrolase (JHEH) is an enzyme that inactivates insect juvenile hormones. This inactivation is accomplished through hydrolysis of the epoxide functional group contained within these hormones into diols. JHEH is one of two enzymes involved in the termination of signaling properties of the various juvenile hormones. The other is juvenile-hormone esterase, or JHE. The first observation of activity from JHEH was in Rhodnius prolixus and Schistocerca gregaria. JH esterase was also first observed in this same work.
- White AF (February 1972). “Metabolism of the juvenile hormone analogue methyl farnesoate 10, 11-epoxide in two insect species”. Life Sciences. 11 (4): 201–210. doi:10.1016/0024-3205(72)90110-5.
Mechanism
JH esterase and epoxide hydrolase are the only known enzymes involved in primary degradation of JH. The inactivated carboxylic acid products of juvenile hormone esterase can be reactivated in peripheral tissues by esterification restoring its biological activity. In contrast, the juvenile hormone diols that are the product of the epoxide hydrolase are very hydrophilic and cannot be converted back to JH. Thus, this enzyme permanently terminates the action of juvenile hormone, unlike JH esterase.
The enzyme mechanism involves the addition of water at the secondary carbon, C-10. In Trichoplusia ni, JH has been found to be the preferred substrate of this enzyme as compared with JH acid.
- Roe RM, Kallapur V, Linderman RJ, Viviani F, Harris SV, Walker EA, Thompson DM (1996). “Mechanism of action and cloning of epoxide hydrolase from the cabbage looper, Trichoplusia ni”. Archives of Insect Biochemistry and Physiology. 32 (3–4): 527–35. doi:10.1002/(SICI)1520-6327(1996)32:3/4<527::AID-ARCH24>3.0.CO;2-D. PMID 8756307.
- Kallapur VL, Majumder C, Roe RM (February 1996). “In vivo and in vitro-tissue specific metabolism of juvenile hormone during the last stadium of the cabbage looper, Trichoplusia ni”. Journal of Insect Physiology. 42 (2): 181–190. doi:10.1016/0022-1910(95)00057-7.
- Lü FG, Fu KY, Guo WC, Li GQ (2015). “Characterization of two juvenile hormone epoxide hydrolases by RNA interference in the Colorado potato beetle”. Gene. 570 (2): 264–71. doi:10.1016/j.gene.2015.06.032. PMID 26079572.
Isoforms
Two forms of JHEH have been cloned from Leptinotarsa decemlineata, which are found in all tissues studied. Knockdown of these enzymes by injected appropriate RNA interference has been found to increase JH titers, prolong larval development, and delay adult emergence, all symptoms of excessive levels of JH.
- Lü FG, Fu KY, Guo WC, Li GQ (2015). “Characterization of two juvenile hormone epoxide hydrolases by RNA interference in the Colorado potato beetle”. Gene. 570 (2): 264–71. doi:10.1016/j.gene.2015.06.032. PMID 26079572.
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents. Two types of small ribonucleic acid (RNA) molecules, microRNA (miRNA) and small interfering RNA (siRNA), are central to components to the RNAi pathway. Once mRNA is degraded, post-transcriptional silencing occurs as protein translation is prevented. Transcription can be inhibited via the pre-transcriptional silencing mechanism of RNAi, through which an enzyme complex catalyzes DNA methylation at genomic positions complementary to complexed siRNA or miRNA. RNAi has an important role in defending cells against parasitic nucleotide sequences (e.g., viruses or transposons) and also influences development of organisms. The RNAi pathway is a naturally occurring process found in many eukaryotes and animal cells. It is initiated by the enzyme Dicer, which cleaves long double-stranded RNA (dsRNA) molecules into short double-stranded fragments of approximately 21 to 23 nucleotide siRNAs. Each siRNA is unwound into two single-stranded RNAs (ssRNAs), the passenger (sense) strand and the guide (antisense) strand. The passenger strand is then cleaved by the protein Argonaute 2 (Ago2). The passenger strand is degraded and the guide strand is incorporated into the RNA-induced silencing complex (RISC). The RISC assembly then binds and degrades the target mRNA. Specifically, this is accomplished when the guide strand pairs with a complementary sequence in a mRNA molecule and induces cleavage by Ago2, a catalytic component of the RISC. In some organisms, this process spreads systemically, despite the initially limited molar concentrations of siRNA. RNAi is a valuable research tool, both in cell culture and in living organisms, because synthetic dsRNA introduced into cells can selectively and robustly induce suppression of specific genes of interest. RNAi may be used for large-scale screens that systematically shut down each gene (and the subsequent proteins it codes for) in the cell, which can help to identify the components necessary for a particular cellular process or an event such as cell division. The pathway is also used as a practical tool for food, medicine and insecticides.
- Saurabh S, Vidyarthi AS, Prasad D (March 2014). “RNA interference: concept to reality in crop improvement”. Planta. 239 (3): 543–64. doi:10.1007/s00425-013-2019-5. PMID 24402564.
- Weiss B, Davidkova G, Zhou LW (March 1999). “Antisense RNA gene therapy for studying and modulating biological processes”. Cellular and Molecular Life Sciences. 55 (3): 334–58. doi:10.1007/s000180050296. PMID 10228554. S2CID 9448271.
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD (November 2005). “Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes”. Cell. 123 (4): 607–20. doi:10.1016/j.cell.2005.08.044. PMID 16271386.
- Kupferschmidt K (August 2013). “A lethal dose of RNA”. Science. 341 (6147): 732–3. Bibcode:2013Sci…341..732K. doi:10.1126/science.341.6147.732. PMID 23950525.
Structure
JHEH is a membrane associated protein, and by photoaffinity labeling has been shown to be a 50 kDA protein in Manduca sexta.
- Touhara K, Soroker V, Prestwich GD (June 1994). “Photoaffinity labeling of juvenile hormone epoxide hydrolase and JH-binding proteins during ovarian and egg development in Manduca sexta”. Insect Biochemistry and Molecular Biology. 24 (6): 633–640. doi:10.1016/0965-1748(94)90100-7.
It has been noted by several that properties of JHEH are similar to those of animal microsomal epoxide hydrolase. Sequence alignments showed that the exact catalytic triad of the animal enzyme (Asp-226, Glu-403 and His-430) is present in JHEH. In addition, the X-ray structure of Bombyx mori JHEH was recently determined.
- Harris SV, Thompson DM, Linderman RJ, Tomalski MD, Roe RM (1999). “Cloning and expression of a novel juvenile hormone-metabolizing epoxide hydrolase during larval-pupal metamorphosis of the cabbage looper, Trichoplusia ni”. Insect Mol. Biol. 8 (1): 85–96. doi:10.1046/j.1365-2583.1999.810085.x. PMID 9927177.
- PDB: 4QLA; Zhou K, Jia N, Hu C, Jiang YL, Yang JP, Chen Y, Li S, Li WF, Zhou CZ (2014). “Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori”. Proteins. 82 (11): 3224–9. doi:10.1002/prot.24676. PMID 25143157.
In enzymology, a microsomal epoxide hydrolase (mEH) (EC 3.3.2.9) is an enzyme that catalyzes the hydrolysis reaction between an epoxide and water to form a diol.
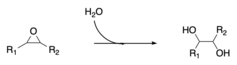
This enzyme plays a role in the uptake of bile salts within the large intestine. It functions as a Na+ dependent transporter. This enzyme participates in metabolism of xenobiotics by cytochrome p450. mEH has been identified as playing a large role in the detoxification and bioactivation of a wide variety of substrates, such as polycyclic aromatic hydrocarbons (PAHs), which are known for their carcinogenic properties. The human homolog of microsomal epoxide hydrolase is EPHX1 and is located on chromosome 1.
- Kiyohara C, Yoshimasu K, Takayama K, Nakanishi Y (January 2006). “EPHX1 polymorphisms and the risk of lung cancer: a HuGE review”. Epidemiology. 17 (1): 89–99. doi:10.1097/01.ede.0000187627.70026.23. PMID 16357600.
- Jackson MR, Craft JA, Burchell B (September 1987). “Nucleotide and deduced amino acid sequence of human liver microsomal epoxide hydrolase”. Nucleic Acids Research. 15 (17): 7188. doi:10.1093/nar/15.17.7188. PMC 306212. PMID 3502697.
A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases). An acid–base–nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence (primary structure). As well as divergent evolution of function (and even the triad’s nucleophile), catalytic triads show some of the best examples of convergent evolution. Chemical constraints on catalysis have led to the same catalytic solution independently evolving in at least 23 separate superfamilies. Their mechanism of action is consequently one of the best studied in biochemistry.
- Dodson G, Wlodawer A (1998). “Catalytic triads and their relatives”. Trends Biochem. Sci. 23 (9): 347–52. doi:10.1016/S0968-0004(98)01254-7. PMID 9787641.
- Buller AR, Townsend CA (2013). “Intrinsic evolutionary constraints on protease structure, enzyme acylation, and the identity of the catalytic triad”. Proc. Natl. Acad. Sci. U.S.A. 110 (8): E653–61. Bibcode:2013PNAS..110E.653B. doi:10.1073/pnas.1221050110. PMC 3581919. PMID 23382230.
- Stryer L, Berg JM, Tymoczko JL (2002). “9 Catalytic Strategies”. Biochemistry (5th ed.). San Francisco: W.H. Freeman. ISBN 9780716749554.
- Perutz M (1992). Protein structure. New approaches to disease and therapy. New York: W.H. Freeman and Co. ISBN 9780716770213.
- Neurath H (1994). “Proteolytic enzymes past and present: the second golden era. Recollections, special section in honor of Max Perutz”. Protein Sci. 3 (10): 1734–9. doi:10.1002/pro.5560031013. PMC 2142620. PMID 7849591.
Post translational modification
Juvenile hormone diol is acted on by juvenile hormone diol kinase, to give juvenile hormone diol phosphate, with the phosphate attached to the hydroxyl group on carbon atom 10. This modification greatly enhances the water solubility, making it easier to excrete.
References
- PDB: 4QLA; Zhou K, Jia N, Hu C, Jiang YL, Yang JP, Chen Y, Li S, Li WF, Zhou CZ (2014). “Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori”. Proteins. 82 (11): 3224–9. doi:10.1002/prot.24676. PMID 25143157.
- White AF (February 1972). “Metabolism of the juvenile hormone analogue methyl farnesoate 10, 11-epoxide in two insect species”. Life Sciences. 11 (4): 201–210. doi:10.1016/0024-3205(72)90110-5.
- Roe RM, Kallapur V, Linderman RJ, Viviani F, Harris SV, Walker EA, Thompson DM (1996). “Mechanism of action and cloning of epoxide hydrolase from the cabbage looper, Trichoplusia ni”. Archives of Insect Biochemistry and Physiology. 32 (3–4): 527–35. doi:10.1002/(SICI)1520-6327(1996)32:3/4<527::AID-ARCH24>3.0.CO;2-D. PMID 8756307.
- Kallapur VL, Majumder C, Roe RM (February 1996). “In vivo and in vitro-tissue specific metabolism of juvenile hormone during the last stadium of the cabbage looper, Trichoplusia ni”. Journal of Insect Physiology. 42 (2): 181–190. doi:10.1016/0022-1910(95)00057-7.
- Lü FG, Fu KY, Guo WC, Li GQ (2015). “Characterization of two juvenile hormone epoxide hydrolases by RNA interference in the Colorado potato beetle”. Gene. 570 (2): 264–71. doi:10.1016/j.gene.2015.06.032. PMID 26079572.
- Touhara K, Soroker V, Prestwich GD (June 1994). “Photoaffinity labeling of juvenile hormone epoxide hydrolase and JH-binding proteins during ovarian and egg development in Manduca sexta”. Insect Biochemistry and Molecular Biology. 24 (6): 633–640. doi:10.1016/0965-1748(94)90100-7.
- Harris SV, Thompson DM, Linderman RJ, Tomalski MD, Roe RM (1999). “Cloning and expression of a novel juvenile hormone-metabolizing epoxide hydrolase during larval-pupal metamorphosis of the cabbage looper, Trichoplusia ni”. Insect Mol. Biol. 8 (1): 85–96. doi:10.1046/j.1365-2583.1999.810085.x. PMID 9927177.
| Hydrolases: ether bond (EC 3.3) |
|---|
Leave a Reply