Von Willebrand factor (VWF) is a blood glycoprotein that promotes hemostasis, specifically, platelet adhesion. It is deficient and/or defective in von Willebrand disease and is involved in many other diseases, including thrombotic thrombocytopenic purpura, Heyde’s syndrome, and possibly hemolytic–uremic syndrome. Increased plasma levels in many cardiovascular, neoplastic, metabolic (e.g. diabetes), and connective tissue diseases are presumed to arise from adverse changes to the endothelium, and may predict an increased risk of thrombosis.
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID 9759493.
- Shahidi M (2017). “Thrombosis and von Willebrand Factor”. Thrombosis and Embolism: From Research to Clinical Practice. Advances in Experimental Medicine and Biology. Vol. 906. pp. 285–306. doi:10.1007/5584_2016_122. ISBN 978-3-319-22107-6. PMID 27628010.
Biochemistry
Synthesis
VWF is a large multimeric glycoprotein present in blood plasma and produced constitutively as ultra-large VWF in endothelium (in the Weibel–Palade bodies), megakaryocytes (α-granules of platelets), and subendothelial connective tissue.
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID 9759493.
Structure
The basic VWF monomer is a 2050-amino acid protein. Every monomer contains a number of specific domains with a specific function; elements of note are:
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID
- the D’/D3 domain, which binds to factor VIII (von Willebrand factor type D domain).
- Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA (July 2012). “Sequence and structure relationships within von Willebrand factor”. Blood. 120 (2): 449–458. doi:10.1182/blood-2012-01-405134. PMC 3398765. PMID 22490677.
- Von Willebrand factor type D domain (vWD) is an evolutionarily-conserved protein domain found in, among others, the von Willebrand factor (vWF). vWF is a large multimeric glycoprotein and it is synthesized by a type of bone marrow cell called megakaryocytes. The vWD domain allows vWF to perform its blood-clotting function by carrying factor VIII around.
- The vWD domain D’/D3 of the von Willebrand factor (vWF) serves as a carrier of clotting factor VIII (FVIII). The native conformation of the D’ domain of vWF is not only required for factor VIII (FVIII) binding but also for normal multimerisation and optimal secretion. The interaction between blood clotting factor VIII and VWF is necessary for normal survival of blood clotting factor VIII in blood circulation. The VWFD domain is a highly structured region, in which the first conserved Cys has been found to form a disulphide bridge with the second conserved one. The other D domains in the protein are necessary for multimerisation.
- Jorieux S, Fressinaud E, Goudemand J, Gaucher C, Meyer D, Mazurier C (May 2000). “Conformational changes in the D’ domain of von Willebrand factor induced by CYS 25 and CYS 95 mutations lead to factor VIII binding defect and multimeric impairment”. Blood. 95 (10): 3139–45. doi:10.1182/blood.V95.10.3139. PMID 10807780.
- This domain is found in mucins, in zonadhesin, in otogelin, and in vitellogenin. Many of these proteins are extracellular glycoproteins. It is also found in a Cypridina-luciferin 2-monooxygenase P17554. Its actual functions in these proteins are unknown.
- Zonadhesin is a protein that in humans is encoded by the ZAN gene. This gene encodes a protein that functions in the species specificity of sperm adhesion to the egg zona pellucida. The encoded protein is located in the acrosome and may be involved in signaling or gamete recognition. An allelic polymorphism in this gene results in both functional and frameshifted alleles; the reference genome represents the functional allele. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Jul 2015].
- GRCh38: Ensembl release 89: ENSG00000146839 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000079173 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Entrez Gene: Zonadhesin (gene/pseudogene)”. Retrieved 2018-06-28.
- Gasper J, Swanson WJ (November 2006). “Molecular population genetics of the gene encoding the human fertilization protein zonadhesin reveals rapid adaptive evolution”. Am. J. Hum. Genet. 79 (5): 820–30. doi:10.1086/508473. PMC 1698559. PMID 17033959.
- Gnanasekar M, Suleman FG, Ramaswamy K, Caldwell JD (October 2009). “Identification of sex hormone binding globulin-interacting proteins in the brain using phage display screening”. Int. J. Mol. Med. 24 (4): 421–6. doi:10.3892/ijmm_00000248. PMID 19724880.
- Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, Casas JP, Sofat R, Kumari M, Rodriguez S, Johnson T, Newhouse SJ, Dominiczak A, Samani NJ, Caulfield M, Sever P, Stanton A, Shields DC, Padmanabhan S, Melander O, Hastie C, Delles C, Ebrahim S, Marmot MG, Smith GD, Lawlor DA, Munroe PB, Day IN, Kivimaki M, Whittaker J, Humphries SE, Hingorani AD (November 2009). “Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip”. Am. J. Hum. Genet. 85 (5): 628–42. doi:10.1016/j.ajhg.2009.10.014. PMC 2775832. PMID 19913121.
- Otogelin is a protein that in humans is encoded by the OTOG gene. The protein encoded by this gene is a component of the acellular membranes of the inner ear. Disruption of the orthologous mouse gene shows that it plays a role in auditory and vestibular functions. It is involved in fibrillar network organization, the anchoring of otoconial membranes and cupulae to the neuroepithelia, and likely in sound stimulation resistance. Mutations in this gene cause autosomal recessive nonsyndromic deafness, type 18B. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, May 2014]. If people don’t have otogelin or otogelin-like they are born with mild or moderate deafness.
- GRCh38: Ensembl release 89: ENSG00000188162 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000009487 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Entrez Gene: Otogelin”. Retrieved 2017-12-12.
- Schraders M, Ruiz-Palmero L, Kalay E, Oostrik J, del Castillo FJ, Sezgin O, Beynon AJ, Strom TM, Pennings RJ, Zazo Seco C, Oonk AM, Kunst HP, Domínguez-Ruiz M, García-Arumi AM, del Campo M, Villamar M, Hoefsloot LH, Moreno F, Admiraal RJ, del Castillo I, Kremer H (2012). “Mutations of the gene encoding otogelin are a cause of autosomal-recessive nonsyndromic moderate hearing impairment”. Am. J. Hum. Genet. 91 (5): 883–9. doi:10.1016/j.ajhg.2012.09.012. PMC 3487128. PMID 23122587.
- Vitellogenin has a couple of mentions in other notes.
- In enzymology, a Cypridina-luciferin 2-monooxygenase (EC 1.13.12.6) is an enzyme that catalyzes the chemical reaction
- Cypridina luciferin + O2 ⇌
oxidized Cypridina luciferin + CO2 + hnu
- Thus, the two substrates of this enzyme are Cypridina luciferin and O2, whereas its 3 products are oxidized Cypridina luciferin, CO2, and light. This enzyme belongs to the family of oxidoreductases, specifically those acting on single donors with O2 as oxidant and incorporation of two atoms of oxygen into the substrate (oxygenases). The oxygen incorporated need not be derived from O with incorporation of one atom of oxygen (internal monooxygenases o internal mixed-function oxidases). The systematic name of this enzyme class is Cypridina-luciferin:oxygen 2-oxidoreductase (decarboxylating). Other names in common use include Cypridina-type luciferase, luciferase (Cypridina luciferin), and Cypridina luciferase. The primary sequence was determined by cloning the cDNA
- Thompson EM, Nagata S, Tsuji FI (1989). “Cloning and expression of cDNA for the luciferase from the marine ostracod Vargula hilgendorfii”. Proceedings of the National Academy of Sciences. 86 (17): 6567–71. doi:10.1073/pnas.86.17.6567. PMC 297885. PMID 2771943.
- Cormier MJ, Crane JM Jr, Nakano Y (1967). “Evidence for the identity of the luminescent systems of Porichthys porosissimus (fish) and Cypridina hilgendorfii (crustacean)”. Biochem. Biophys. Res. Commun. 29 (5): 747–52. doi:10.1016/0006-291X(67)90281-1. PMID 5624784.
- Karpetsky TP, White EH (1973). “The synthesis of Cypridina etioluciferamine and the proof of the structure of Cypridina luciferin”. Tetrahedron. 29 (23): 3761–3773. doi:10.1016/0040-4020(73)80193-0.
- Kishi Y, Goto T, Hirata Y, Shiromura O, Johnson FH (1966). “Cypridina bioluminescence. I. Structure of Cypridina luciferin”. Tetrahedron Lett. 7: 3427–3436. doi:10.1016/S0040-4039(01)82806-9.
- Tsuji FI, Lynch RV III, Stevens CL (1974). “Some properties of luciferase from the bioluminescent crustacean, Cypridina hilgendorfii”. Biochemistry. 13 (25): 5204–9. doi:10.1021/bi00722a024. PMID 4433517.
- Cypridina luciferin + O2 ⇌
- Zonadhesin is a protein that in humans is encoded by the ZAN gene. This gene encodes a protein that functions in the species specificity of sperm adhesion to the egg zona pellucida. The encoded protein is located in the acrosome and may be involved in signaling or gamete recognition. An allelic polymorphism in this gene results in both functional and frameshifted alleles; the reference genome represents the functional allele. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Jul 2015].
- Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA (July 2012). “Sequence and structure relationships within von Willebrand factor”. Blood. 120 (2): 449–458. doi:10.1182/blood-2012-01-405134. PMC 3398765. PMID 22490677.
- the A1 domain, which binds to:
- the A2 domain, which must partially unfold to expose the buried cleavage site for the specific ADAMTS13 protease that inactivates VWF by making much smaller multimers. The partial unfolding is affected by shear flow in the blood, by calcium binding, and by the lump of a sequence-adjacent “vicinal disulfide” at the A2-domain C-terminus.
- Jakobi AJ, Mashaghi A, Tans SJ, Huizinga EG (July 2011). “Calcium modulates force sensing by the von Willebrand factor A2 domain”. Nature Communications. 2: 385. Bibcode:2011NatCo…2..385J. doi:10.1038/ncomms1385. PMC 3144584. PMID 21750539.
- Luken BM, Winn LY, Emsley J, Lane DA, Crawley JT (June 2010). “The importance of vicinal cysteines, C1669 and C1670, for von Willebrand factor A2 domain function”. Blood. 115 (23): 4910–4913. doi:10.1182/blood-2009-12-257949. PMC 2890177. PMID 20354169.
- the A3 domain, which binds to collagen (von Willebrand factor type A domain)
- The von Willebrand factor type A (vWA) domain is a protein domain named after its occurrence in von Willebrand factor (vWF), a large multimeric glycoprotein found in blood plasma. Mutant forms of vWF are involved in the aetiology of bleeding disorders. This type A domain is the prototype for a protein superfamily (InterPro: IPR036465; see also Pfam clan).
- Qu A, Leahy DJ (October 1995). “Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, alpha L beta 2) integrin”. Proc. Natl. Acad. Sci. U.S.A. 92 (22): 10277–81. doi:10.1073/pnas.92.22.10277. PMC 40779. PMID 7479767.
- Ruggeri ZM, Ware J (1993). “von Willebrand factor”. FASEB J. 7 (2): 308–316. doi:10.1096/fasebj.7.2.8440408. PMID 8440408. S2CID 10574567.
- The vWA domain is found in various plasma proteins: complement factors B, C2, CR3 and CR4; the integrins (I-domains); collagen types VI, VII, XII and XIV; and other extracellular proteins. Although the majority of vWA-containing proteins are extracellular, the most ancient ones present in all eukaryotes are all intracellular proteins involved in functions such as transcription, DNA repair, ribosomal and membrane transport and the proteasome. A common feature appears to be involvement in multiprotein complexes. Proteins that incorporate vWA domains participate in numerous biological events (e.g. cell adhesion, migration, homing, pattern formation, and signal transduction), involving interaction with a large array of ligands. A number of human diseases arise from mutations in vWA domains.
- Colombatti A, Bonaldo P, Doliana R (1993). “Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins”. Matrix. 13 (4): 297–306. doi:10.1016/S0934-8832(11)80025-9. PMID 8412987.
- Smith KF, Haris PI, Chapman D, Perkins SJ, Williams SC, Sim RB (1994). “The secondary structure of the von Willebrand factor type A domain in factor B of human complement by Fourier transform infrared spectroscopy. Its occurrence in collagen types VI, VII, XII and XIV, the integrins and other proteins by averaged structure predictions”. J. Mol. Biol. 238 (1): 104–119. doi:10.1006/jmbi.1994.1271. PMID 8145250.
- Bork P (1991). “Shuffled domains in extracellular proteins”. FEBS Lett. 286 (1): 47–54. doi:10.1016/0014-5793(91)80937-X. PMID 1864378. S2CID 22126481.
- Secondary structure prediction from 75 aligned vWA sequences has revealed a largely alternating sequence of alpha-helices and beta-strands. Fold recognition algorithms were used to score sequence compatibility with a library of known structures: the vWA domain fold was predicted to be a doubly wound, open, twisted beta-sheet flanked by alpha-helices. 3D structures have been determined for the I-domains of integrins CD11b (with bound magnesium) and CD11a (with bound manganese). The domain adopts a classic alpha/beta Rossmann fold and contains an unusual metal ion coordination site at its surface. It has been suggested that this site represents a general metal ion-dependent adhesion site (MIDAS) for binding protein ligands. The residues constituting the MIDAS motif in the CD11b and CD11a I-domains are completely conserved, but the manner in which the metal ion is coordinated differs slightly.
- Smith KF, Haris PI, Chapman D, Perkins SJ, Williams SC, Sim RB (1994). “The secondary structure of the von Willebrand factor type A domain in factor B of human complement by Fourier transform infrared spectroscopy. Its occurrence in collagen types VI, VII, XII and XIV, the integrins and other proteins by averaged structure predictions”. J. Mol. Biol. 238 (1): 104–119. doi:10.1006/jmbi.1994.1271. PMID 8145250.
- Bork P (1991). “Shuffled domains in extracellular proteins”. FEBS Lett. 286 (1): 47–54. doi:10.1016/0014-5793(91)80937-X. PMID 1864378. S2CID 22126481.
- Perkins SJ, Edwards YJ (1995). “The protein fold of the von Willebrand factor type A domain is predicted to be similar to the open twisted beta-sheet flanked by alpha-helices found in human ras-p21”. FEBS Lett. 358 (3): 283–286. doi:10.1016/0014-5793(94)01447-9. PMID 7843416. S2CID 85317759.
- Lee JO, Rieu P, Arnaout MA, Liddington R (1995). “Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18)”. Cell. 80 (4): 631–638. doi:10.1016/0092-8674(95)90517-0. PMID 7867070. S2CID 11275359.
- Leahy DJ, Qu A (1995). “Crystal structure of the I-domain from the CD11a/CD18 (LFA-1, alpha L beta 2) integrin”. Proc. Natl. Acad. Sci. U.S.A. 92 (22): 10277–10281. doi:10.1073/pnas.92.22.10277. PMC 40779. PMID 7479767.
- Bork, P; Rohde, K (1991). “More von Willebrand factor type a domains? Sequence similarities with malaria thrombospondin-related anonymous protein, dihydropyridine-sensitive calcium channel and inter-alpha-trypsin inhibitor”. The Biochemical Journal. 279 (3): 908–10. doi:10.1042/bj2790908. PMC 1151534. PMID 1659389.
- Human proteins containing this domain
- ANTXR1; ANTXR2; BF; C2; CACHD1; CACNA2D1; CACNA2D2; CACNA2D3; CACNA2D4; CFB; CLCA1; CLCA2; CLCA4; COCH; COL12A1; COL14A1; COL20A1; COL21A1; COL22A1; COL28; COL6A1; COL6A2; COL6A3; COL7A1; COLA1L; CaCC1; ITGA1; ITGA10; ITGA11; ITGA2; ITGAD; ITGAE; ITGAL; ITGAM; ITGAX; ITIH1; ITIH2; ITIH3; ITIH4; ITIH5; ITIH5L; LOC285929; LOC340267; LOC389462; LOH11CR2A; MATN1; MATN2; MATN3; MATN4; PARP4; SVEP1(SEL-OB); VIT; VWA1; VWA2; VWF; hCLCA1; hCLCA2; CMG2;
- The von Willebrand factor type A (vWA) domain is a protein domain named after its occurrence in von Willebrand factor (vWF), a large multimeric glycoprotein found in blood plasma. Mutant forms of vWF are involved in the aetiology of bleeding disorders. This type A domain is the prototype for a protein superfamily (InterPro: IPR036465; see also Pfam clan).
- the C4 domain, in which the RGD motif binds to platelet integrin αIIbβ3 when this is activated (von Willebrand factor type C domain)
- Von Willebrand factor, type C (VWFC or VWC)is a protein domain is found in various blood plasma proteins: complement factors B, C2, CR3 and CR4; the integrins (I-domains); collagen types VI, VII, XII and XIV; and other extracellular proteins. Although the majority of VWA-containing proteins are extracellular, the most ancient ones present in all eukaryotes are all intracellular proteins involved in functions such as transcription, DNA repair, ribosomal and membrane transport and the proteasome. A common feature appears to be involvement in multiprotein complexes. Proteins that incorporate vWF domains participate in numerous biological events (e.g. cell adhesion, migration, homing, pattern formation, and signal transduction), involving interaction with a large array of ligands.
- Colombatti A, Bonaldo P, Doliana R (1993). “Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins”. Matrix. 13 (4): 297–306. doi:10.1016/S0934-8832(11)80025-9. PMID 8412987.
- Smith KF, Haris PI, Chapman D, Perkins SJ, Williams SC, Sim RB (1994). “The secondary structure of the von Willebrand factor type A domain in factor B of human complement by Fourier transform infrared spectroscopy. Its occurrence in collagen types VI, VII, XII and XIV, the integrins and other proteins by averaged structure predictions”. J. Mol. Biol. 238 (1): 104–119. doi:10.1006/jmbi.1994.1271. PMID 8145250.
- Bork P (1991). “Shuffled domains in extracellular proteins”. FEBS Lett. 286 (1): 47–54. doi:10.1016/0014-5793(91)80937-X. PMID 1864378. S2CID 22126481.
- A number of human diseases arise from mutations in VWA domains. The domain is named after the von Willebrand factor (VWF) type C repeat which is found in multidomain protein/multifunctional proteins involved in maintaining homeostasis. For the von Willebrand factor the duplicated VWFC domain is thought to participate in oligomerization, but not in the initial dimerization step. The presence of this region in a number of other complex-forming proteins points to the possible involvement of the VWFC domain in complex formation.
- Smith KF, Haris PI, Chapman D, Perkins SJ, Williams SC, Sim RB (1994). “The secondary structure of the von Willebrand factor type A domain in factor B of human complement by Fourier transform infrared spectroscopy. Its occurrence in collagen types VI, VII, XII and XIV, the integrins and other proteins by averaged structure predictions”. J. Mol. Biol. 238 (1): 104–119. doi:10.1006/jmbi.1994.1271. PMID 8145250.
- Bork P (1991). “Shuffled domains in extracellular proteins”. FEBS Lett. 286 (1): 47–54. doi:10.1016/0014-5793(91)80937-X. PMID 1864378. S2CID 22126481.
- Hunt LT, Barker WC (1987). “von Willebrand factor shares a distinctive cysteine-rich domain with thrombospondin and procollagen”. Biochem. Biophys. Res. Commun. 144 (2): 876–882. doi:10.1016/S0006-291X(87)80046-3. PMID 3495268.
- Voorberg J, Fontijn R, Calafat J, Janssen H, van Mourik JA, Pannekoek H (1991). “Assembly and routing of von Willebrand factor variants: the requirements for disulfide-linked dimerization reside within the carboxy-terminal 151 amino acids”. J. Cell Biol. 113 (1): 195–205. doi:10.1083/jcb.113.1.195. PMC 2288914. PMID 2007623.
- Human proteins containing this domain
- BMP binding endothelial regulator (BMPER)
- Cysteine-rich motor neuron 1 protein (CRIM1)
- Extracellular matrix protein 2 (ECM2)
- Fraser extracellular matrix complex subunit 1 (FRAS1)
- Neural EGFL like 1 (NELL1)
- Neural EGFL like 2 (NELL2)
- Peroxidasin like (PXDNL)
- Von Willebrand factor C and EGF domain-containing protein (VWCE)
- Von Willebrand factor (VWF)
- Chordin family
- Chordin (CHRD)
- Chordin-like 1 (CHRDL1)
- Chordin-like 2 (CHRDL2)
- Brorin (VWC2)
- Collagen family
- Collagen, type I, alpha 1 (COL1A1)
- Collagen, type II, alpha 1 (COL2A1)
- Collagen, type III, alpha 1 (COL3A1)
- Collagen, type V, alpha 2 (COL5A2)
- Mucin family
- Thrombospondin superfamily
- Thrombospondin 1 (THBS1)
- Thrombospondin 2 (THBS2)
- SCO-spondin (SSPO)
- CCN family
- CCN1: Cysteine-rich angiogenic inducer 61 (CYR61)
- CCN2: Connective tissue growth factor (CTGF)
- CCN3: Nephroblastoma overexpressed (NOV)
- CCN4: WNT1-inducible-signaling pathway protein 1 (WISP1)
- CCN5: WNT1-inducible-signaling pathway protein 2 (WISP2)
- Von Willebrand factor, type C (VWFC or VWC)is a protein domain is found in various blood plasma proteins: complement factors B, C2, CR3 and CR4; the integrins (I-domains); collagen types VI, VII, XII and XIV; and other extracellular proteins. Although the majority of VWA-containing proteins are extracellular, the most ancient ones present in all eukaryotes are all intracellular proteins involved in functions such as transcription, DNA repair, ribosomal and membrane transport and the proteasome. A common feature appears to be involvement in multiprotein complexes. Proteins that incorporate vWF domains participate in numerous biological events (e.g. cell adhesion, migration, homing, pattern formation, and signal transduction), involving interaction with a large array of ligands.
- the other C domains, which may interact in ER dimers: the larger protein show six beads of (C and C-like) domains under cryo-EM.
- Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA (July 2012). “Sequence and structure relationships within von Willebrand factor”. Blood. 120 (2): 449–458. doi:10.1182/blood-2012-01-405134. PMC 3398765. PMID 22490677.
- the “cystine knot” domain (at the C-terminal end of the protein), which VWF shares with platelet-derived growth factor (PDGF), transforming growth factor-β (TGFβ) and β-human chorionic gonadotropin (βHCG, of pregnancy test fame). (von Willebrand factor type C domain)
- A cystine knot is a protein structural motif containing three disulfide bridges (formed from pairs of cysteine residues). The sections of polypeptide that occur between two of them form a loop through which a third disulfide bond passes, forming a rotaxane substructure. The cystine knot motif stabilizes protein structure and is conserved in proteins across various species.
- Wu H, Lustbader JW, Liu Y, Canfield RE, Hendrickson WA (June 1994). “Structure of human chorionic gonadotropin at 2.6 A resolution from MAD analysis of the selenomethionyl protein”. Structure. 2 (6): 545–58. doi:10.1016/s0969-2126(00)00054-x. PMID 7922031.
- “Cystine Knots”. The Cyclotide Webpage. Archived from the original on 2015-02-05. Retrieved 2019-04-24.
- Sherbet, G.V. (2011), “Growth Factor Families”, Growth Factors and Their Receptors in Cell Differentiation, Cancer and Cancer Therapy, Elsevier, pp. 3–5, doi:10.1016/b978-0-12-387819-9.00002-5, ISBN 9780123878199, retrieved 2019-05-01
- Vitt, Ursula A.; Hsu, Sheau Y.; Hsueh, Aaron J. W. (2001-05-01). “Evolution and Classification of Cystine Knot-Containing Hormones and Related Extracellular Signaling Molecules”. Molecular Endocrinology. 15 (5): 681–694. doi:10.1210/mend.15.5.0639. ISSN 0888-8809. PMID 11328851.
- Daly NL, Craik DJ (June 2011). “Bioactive cystine knot proteins”. Current Opinion in Chemical Biology. 15 (3): 362–8. doi:10.1016/j.cbpa.2011.02.008. PMID 21362584.
- There are three types of cystine knot, which differ in the topology of the disulfide bonds:
- The growth factor cystine knot (GFCK)
- inhibitor cystine knot (ICK) common in spider and snail toxins
- Cyclic Cystine Knot, or cyclotide
- The growth factor cystine knot was first observed in the structure of nerve growth factor (NGF), solved by X-ray crystallography and published in 1991 by Tom Blundell in Nature. The GFCK is present in four superfamilies. These include nerve growth factor, transforming growth factor beta (TGF-β), platelet-derived growth factor, and glycoprotein hormones including human chorionic gonadotropin. These are structurally related due to the presence of the cystine knot motif but differ in sequence. All GFCK structures that have been determined are dimeric, but their dimerization modes in different classes are different. The vascular endothelial growth factor subfamily, categorized as part of the platelet-derived growth factor superfamily, includes proteins that are angiogenic factors.
- PDB: 1bet; McDonald NQ, Lapatto R, Murray-Rust J, Gunning J, Wlodawer A, Blundell TL (December 1991). “New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor”. Nature. 354 (6352): 411–4. Bibcode:1991Natur.354..411M. doi:10.1038/354411a0. PMID 1956407. S2CID 4346788.
- Sun PD, Davies DR (1995). “The cystine-knot growth-factor superfamily”. Annual Review of Biophysics and Biomolecular Structure. 24 (1): 269–91. doi:10.1146/annurev.bb.24.060195.001413. PMID 7663117.
- Jiang X, Dias JA, He X (January 2014). “Structural biology of glycoprotein hormones and their receptors: insights to signaling”. Molecular and Cellular Endocrinology. 382 (1): 424–451. doi:10.1016/j.mce.2013.08.021. PMID 24001578.
- Iyer S, Acharya KR (November 2011). “Tying the knot: the cystine signature and molecular-recognition processes of the vascular endothelial growth factor family of angiogenic cytokines”. The FEBS Journal. 278 (22): 4304–22. doi:10.1111/j.1742-4658.2011.08350.x. PMC 3328748. PMID 21917115.
- The presence of the cyclic cystine knot (CCK) motif was discovered when cyclotides were isolated from various plant families. The CCK motif has a cyclic backbone, triple stranded beta sheet, and cystine knot conformation.
- Craik DJ, Daly NL, Bond T, Waine C (December 1999). “Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif”. Journal of Molecular Biology. 294 (5): 1327–36. doi:10.1006/jmbi.1999.3383. PMID 10600388.
- Novel proteins are being added to the cystine knot motif family, also known as the C-terminal cystine knot (CTCK) proteins. They share approximately 90 amino acid residues in their cysteine-rich C-terminal regions.
- Iyer S, Acharya KR (November 2011). “Tying the knot: the cystine signature and molecular-recognition processes of the vascular endothelial growth factor family of angiogenic cytokines”. The FEBS Journal. 278 (22): 4304–22. doi:10.1111/j.1742-4658.2011.08350.x. PMC 3328748. PMID 21917115.
- Inhibitor cystine knot (ICK) is a structural motif with a triple stranded antiparallel beta sheet linked by three disulfide bonds, forming a knotted core. The ICK motif can be found under the category of phylum, such as animals and plants. It is often found in many venom peptides such as those of snails, spiders, and scorpions. Peptide K-PVIIA, which contains an ICK, can undergo a successful enzymatic backbone cyclization. The disulfide connectivity and the common sequence pattern of the ICK motif provides the stability of the peptides that support cyclization.
- Kwon, Soohyun; Bosmans, Frank; Kaas, Quentin; Cheneval, Oliver; Cinibear, Anne C; Rosengren, K Johan; Wang, Conan K; Schroeder, Christina I; Craik, David J (19 April 2016). “Efficient enzymatic cyclization of an inhibitory cystine knot‐containing peptide”. Biotechnology and Bioengineering. 113 (10): 2202–2212. doi:10.1002/bit.25993. PMC 5526200. PMID 27093300.
- The stability and structure of the cystine knot motif implicates possible applications in drug design. The hydrogen bonding between the disulfide bonds of the motif and beta-sheet structures gives rise to highly efficient structure stabilization. In addition, the size of the motif is approximately 30 amino acid residues. These two characteristics make it an attractive biomolecule to be used for drug delivery as it exhibits thermal stability, chemical stability, and proteolytic resistance. The biological activities of these molecules are partially due to the unique interlocking arrangement and cyclized peptide backbone which contains a conserved sequence shared among circulins. Circulins have previously been identified in a screen for anti-HIV activity. Studies have shown that cystine knot proteins can be incubated at temperatures of 65 °C or placed in 1N HCl/1N NaOH without loss of structural and functional integrity. Its resistance from oral and some intestinal proteases suggest possible use for oral delivery. Possible future applications include pain relief as well as antiviral and antibacterial functions.
- Kolmar, Harald. “Biological Diversity and Therapeutic Potential of Natural and Engineered Cystine Knot Miniproteins.” Current Opinion in Pharmacology, vol. 9, no. 5, 2009, pp. 608–614., doi:10.1016/j.coph.2009.05.004.
- K.R. Gustafson, R.C. Sowder II, L.E. Henderson, I.C. Parsons, Y. Kashman, J.H. Cardellina II, J.B. McMahon, R.W. Buckheit Jr., L.K. Pannell, M.R. Boyd Circulins A and B: novel HIV-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia J. Am. Chem. Soc., 116 (1994), pp. 9337-9338
- Craik, David J., et al. “The Cystine Knot Motif in Toxins and Implications for Drug Design.” Toxicon, vol. 39, no. 1, 2001, pp. 43–60., doi:10.1016/s0041-0101(00)00160-4.
- A cystine knot is a protein structural motif containing three disulfide bridges (formed from pairs of cysteine residues). The sections of polypeptide that occur between two of them form a loop through which a third disulfide bond passes, forming a rotaxane substructure. The cystine knot motif stabilizes protein structure and is conserved in proteins across various species.
Monomers are subsequently N-glycosylated, arranged into dimers in the endoplasmic reticulum and into multimers in the Golgi apparatus by crosslinking of cysteine residues via disulfide bonds. With respect to the glycosylation, VWF is one of only a few proteins that carry ABO blood group system antigens. VWFs coming out of the Golgi are packaged into storage organelles, Weibel-Palade bodies (WPBs) in endothelial cells and α-granules in platelets.
- Lenting PJ, Christophe OD, Denis CV (March 2015). “von Willebrand factor biosynthesis, secretion, and clearance: connecting the far ends”. Blood. 125 (13): 2019–2028. doi:10.1182/blood-2014-06-528406. PMID 25712991. S2CID 27785232.
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID 9759493.
Multimers of VWF can be extremely large, >20,000 kDa, and consist of over 80 subunits of 250 kDa each. Only the large multimers are functional. Some cleavage products that result from VWF production are also secreted but probably serve no function.
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID 9759493.
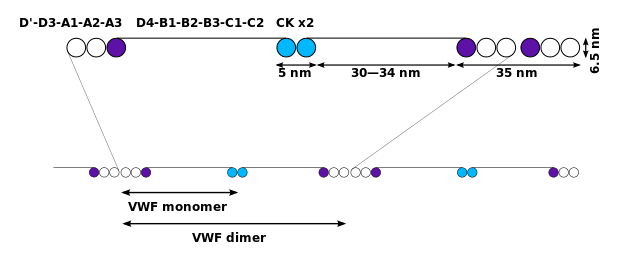
Function
Von Willebrand Factor’s primary function is binding to other proteins, in particular factor VIII, and it is important in platelet adhesion to wound sites. It is not an enzyme and, thus, has no catalytic activity.
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID 9759493.
VWF binds to a number of cells and molecules. The most important ones are:
- Factor VIII is bound to VWF while inactive in circulation; factor VIII degrades rapidly when not bound to VWF. Factor VIII is released from VWF by the action of thrombin. In the absence of VWF, factor VIII has a half-life of 1–2 hours; when carried by intact VWF, factor VIII has a half-life of 8–12 hours.
- VWF binds to collagen, e.g., when collagen is exposed beneath endothelial cells due to damage occurring to the blood vessel. Endothelium also releases VWF which forms additional links between the platelets’ glycoprotein Ib/IX/V and the collagen fibrils
- VWF binds to platelet gpIb when it forms a complex with gpIX and gpV;
- Glycoprotein Ib (GPIb), also known as CD42, is a component of the GPIb-V-IX complex on platelets. The GPIb-V-IX complex binds von Willebrand factor, allowing platelet adhesion and platelet plug formation at sites of vascular injury. Glycoprotein Ibα (GPIbα) is the major ligand-binding subunit of the GPIb-V-IX complex. GPIbα is heavily glycosylated. It is deficient in the Bernard–Soulier syndrome. A gain-of-function mutation causes platelet-type von Willebrand disease. Autoantibodies against Ib/IX can be produced in immune thrombocytopenic purpura. Components include GP1BA and GP1BB. It complexes with Glycoprotein IX.
- Bode AP, Read MS, Reddick RL (February 1999). “Activation and adherence of lyophilized human platelets on canine vessel strips in the Baumgartner perfusion chamber”. The Journal of Laboratory and Clinical Medicine. 133 (2): 200–211. doi:10.1016/S0022-2143(99)90013-6. PMID 9989772.
- Hollenhorst MA, Tiemeyer KH, Mahoney KE, Aoki K, Ishihara M, Lowery SC, et al. (April 2023). “Comprehensive analysis of platelet glycoprotein Ibα ectodomain glycosylation”. Journal of Thrombosis and Haemostasis. 21 (4): 995–1009. doi:10.1016/j.jtha.2023.01.009. PMC 10065957. PMID 36740532.
- McPherson RA, Pincus MR (2007). Henry’s Clinical Diagnosis and Management by Laboratory Methods (21st ed.). Philadelphia, Pa: Saunders Elsevier. pp. 760–762. ISBN 978-1-4160-0287-1.
- McMillan R (October 2007). “The pathogenesis of chronic immune thrombocytopenic purpura”. Seminars in Hematology. 44 (4 Suppl 5): S3–S11. doi:10.1053/j.seminhematol.2007.11.002. PMID 18096470.
- Glycoprotein IX (platelet) (GP9) also known as CD42a (Cluster of Differentiation 42a), is a human gene. Platelet glycoprotein IX (GP9) is a small membrane glycoprotein found on the surface of human platelets. It forms a 1-to-1 noncovalent complex with glycoprotein Ib (GP Ib), a platelet surface membrane glycoprotein complex that functions as a receptor for von Willebrand factor (VWF; MIM 193400) (known as the Glycoprotein Ib-IX-V Receptor Complex). The main portion of the receptor is a heterodimer composed of 2 polypeptide chains, an alpha chain (GP1BA; MIM 606672) and a beta chain (GP1BB; MIM 138720), that are linked by disulfide bonds. The complete receptor complex includes noncovalent association of the alpha and beta subunits with GP9 and platelet glycoprotein V (GP5; MIM 173511).[supplied by OMIM] See also Cluster of differentiation
- GRCh38: Ensembl release 89: ENSG00000169704 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000030054 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Entrez Gene: GP9 glycoprotein IX (platelet)”.
- Factor V (pronounced factor five) is a protein of the coagulation system, rarely referred to as proaccelerin or labile factor. In contrast to most other coagulation factors, it is not enzymatically active but functions as a cofactor. Deficiency leads to predisposition for hemorrhage, while some mutations (most notably factor V Leiden) predispose for thrombosis.
- “F5 gene: MedlinePlus Genetics”. medlineplus.gov. Retrieved 2023-03-25.
- The gene for factor V is located on the first chromosome (1q24). It is genomically related to the family of multicopper oxidases, and is homologous to coagulation factor VIII. The gene spans 70 kb, consists of 25 exons, and the resulting protein has a relative molecular mass of approximately 330kDa. Factor V protein consists of six domains: A1-A2-B-A3-C1-C2. The A domains are homologous to the A domains of the copper-binding protein ceruloplasmin, and form a triangular as in that protein. A copper ion is bound in the A1-A3 interface, and A3 interacts with the plasma. The C domains belong to the phospholipid-binding discoidin domain family (unrelated to C2 domain), and the C2 domain mediates membrane binding. The B domain C-terminus acts as a cofactor for the anticoagulant protein C activation by protein S. Activation of factor V to factor Va is done by cleavage and release of the B domain, after which the protein no longer assists in activating protein C. The protein is now divided to a heavy chain, consisting of the A1-A2 domains, and a light chain, consisting of the A3-C1-C2 domains. Both form non-covalently a complex in a calcium-dependent manner. This complex is the pro-coagulant factor Va.
- Villoutreix BO, Dahlbäck B (June 1998). “Structural investigation of the A domains of human blood coagulation factor V by molecular modeling”. Protein Science. 7 (6): 1317–25. doi:10.1002/pro.5560070607. PMC 2144041. PMID 9655335.
- Thorelli E, Kaufman RJ, Dahlbäck B (June 1998). “The C-terminal region of the factor V B-domain is crucial for the anticoagulant activity of factor V”. The Journal of Biological Chemistry. 273 (26): 16140–45. doi:10.1074/jbc.273.26.16140. PMID 9632668.
- Macedo-Ribeiro S, Bode W, Huber R, Quinn-Allen MA, Kim SW, Ortel TL, Bourenkov GP, Bartunik HD, Stubbs MT, Kane WH, Fuentes-Prior P (November 1999). “Crystal structures of the membrane-binding C2 domain of human coagulation factor V”. Nature. 402 (6760): 434–39. Bibcode:1999Natur.402..434M. doi:10.1038/46594. PMID 10586886. S2CID 4393638.
- Factor V synthesis occurs in the liver, principally. In the liver, it is primarily produced by megakaryocytes, which produce platelets and platelet-derived factor V, and hepatocytes, which produce plasma-derived factor V. The molecule circulates in plasma as a single-chain molecule with a plasma half-life of 12–36 hours. Factor V is able to bind to activated platelets and is activated by thrombin. On activation, factor V is spliced in two chains (heavy and light chain with molecular masses of 110000 and 73000, respectively) which are noncovalently bound to each other by calcium. The thereby activated factor V (now called FVa) is a cofactor of the prothrombinase complex: The activated factor X (FXa) enzyme requires calcium and activated factor V (FVa) to convert prothrombin to thrombin on the cell surface membrane. Factor Va is degraded by activated protein C, one of the principal physiological inhibitors of coagulation. In the presence of thrombomodulin, thrombin acts to decrease clotting by activating protein C; therefore, the concentration and action of protein C are important determinants in the negative feedback loop through which thrombin limits its own activation.
- Lam W, Moosavi L (18 July 2022). “Physiology, Factor V”. StatPearls. StatPearls Publishing. PMID 31334957. Retrieved 7 February 2022.
- Huang JN, Koerper MA (November 2008). “Factor V deficiency: a concise review”. Haemophilia. 14 (6): 1164–69. doi:10.1111/j.1365-2516.2008.01785.x. PMID 19141156.
- Various hereditary disorders of factor V are known. Deficiency is associated with a rare mild form of hemophilia (termed parahemophilia or Owren parahemophilia), the incidence of which is about 1:1,000,000. It inherits in an autosomal recessive fashion. There exists a bleeding tendency associated with the genetic up‐regulation of FV‐short, a minor splicing isoform of FV. This abnormal bleeding tendency occurs in east Texas bleeding disorder, Amsterdam bleeding disorder, and a third and more extreme example described in 2021 by Karen L. Zimowski et al. Other mutations of factor V are associated with venous thrombosis. They are the most common hereditary causes for thrombophilia (a tendency to form blood clots). The most common one of these, factor V Leiden, is due to the replacement of an arginine residue with glutamine at amino acid position 506 (R506Q). All prothrombotic factor V mutations (factor V Leiden, factor V Cambridge, factor V Hong Kong) make it resistant to cleavage by activated protein C (“APC resistance”). It therefore remains active and increases the rate of thrombin generation.
- Castoldi E (July 2021). “F5-Atlanta: Factor V-short strikes again”. Journal of Thrombosis and Haemostasis. 19 (7): 1638–1640. doi:10.1111/jth.15351. PMC 8362210. PMID 34176223.
- The possibility of an extra coagulation factor was initially resisted on methodological grounds by Drs Armand Quick and Walter Seegers, both world authorities in coagulation. Confirmatory studies from other groups led to their final approval several years later. Owren initially felt that factor V (labile factor or proaccelerin) activated another factor, which he named VI. VI was the factor that accelerated the conversion from prothrombin to thrombin. It was later discovered that factor V was “converted” (activated) by thrombin itself, and later still that factor VI was simply the activated form of factor V. The complete amino acid sequence of the protein was published in 1987. In 1994 factor V Leiden, resistant to inactivation by protein C, was described; this abnormality is the most common genetic cause for thrombosis.
- Stormorken H (February 2003). “The discovery of factor V: a tricky clotting factor”. Journal of Thrombosis and Haemostasis. 1 (2): 206–13. doi:10.1046/j.1538-7836.2003.00043.x. PMID 12871488.
- Owren PA (April 1947). “Parahaemophilia; haemorrhagic diathesis due to absence of a previously unknown clotting factor”. Lancet. 1 (6449): 446–48. doi:10.1016/S0140-6736(47)91941-7. PMID 20293060.
- Jenny RJ, Pittman DD, Toole JJ, Kriz RW, Aldape RA, Hewick RM, Kaufman RJ, Mann KG (July 1987). “Complete cDNA and derived amino acid sequence of human factor V”. Proceedings of the National Academy of Sciences of the United States of America. 84 (14): 4846–50. Bibcode:1987PNAS…84.4846J. doi:10.1073/pnas.84.14.4846. PMC 305202. PMID 3110773.
- Bertina RM, Koeleman BP, Koster T, Rosendaal FR, Dirven RJ, de Ronde H, van der Velden PA, Reitsma PH (May 1994). “Mutation in blood coagulation factor V associated with resistance to activated protein C”. Nature. 369 (6475): 64–67. Bibcode:1994Natur.369…64B. doi:10.1038/369064a0. PMID 8164741. S2CID 4314040.
- Factor V has been shown to interact with Protein S.
- Heeb MJ, Kojima Y, Rosing J, Tans G, Griffin JH (December 1999). “C-terminal residues 621–635 of protein S are essential for binding to factor Va”. The Journal of Biological Chemistry. 274 (51): 36187–92. doi:10.1074/jbc.274.51.36187. PMID 10593904.
- Heeb MJ, Mesters RM, Tans G, Rosing J, Griffin JH (February 1993). “Binding of protein S to factor Va associated with inhibition of prothrombinase that is independent of activated protein C”. The Journal of Biological Chemistry. 268 (4): 2872–77. doi:10.1016/S0021-9258(18)53854-0. PMID 8428962.
- Protein S (also known as PROS) is a vitamin K-dependent plasma glycoprotein synthesized in the liver. In the circulation, Protein S exists in two forms: a free form and a complex form bound to complement protein C4b-binding protein (C4BP). In humans, protein S is encoded by the PROS1 gene. Protein S plays a role in coagulation. Protein S is named for Seattle, Washington, where it was originally discovered and purified by Earl Davie‘s group in 1977.
- Lundwall A, Dackowski W, Cohen E, Shaffer M, Mahr A, Dahlbäck B, Stenflo J, Wydro R (September 1986). “Isolation and sequence of the cDNA for human protein S, a regulator of blood coagulation”. Proc. Natl. Acad. Sci. U.S.A. 83 (18): 6716–20. Bibcode:1986PNAS…83.6716L. doi:10.1073/pnas.83.18.6716. PMC 386580. PMID 2944113.
- Long GL, Marshall A, Gardner JC, Naylor SL (January 1988). “Genes for human vitamin K-dependent plasma proteins C and S are located on chromosomes 2 and 3, respectively”. Somat. Cell Mol. Genet. 14 (1): 93–8. doi:10.1007/BF01535052. PMID 2829367. S2CID 31236887.
- “Protein S deficiency”. UpToDate. Retrieved May 10, 2017.
- Kaushansky K, Lichtman M, Prchal J, Levi M, Press O, Burns L, Caligiuri M (2015). Williams Hematology. McGraw-Hill. p. 1926.
- Protein S is partly homologous to other vitamin K-dependent plasma coagulation proteins, such as protein C and factors VII, IX, and X. Similar to them, it has a Gla domain and several EGF-like domains (four rather than two), but no serine protease domain. Instead, there is a large C-terminus domain that is homologous to plasma steroid hormone-binding proteins such as sex hormone-binding globulin and corticosteroid-binding globulin. It may play a role in the protein functions as either a cofactor for activated protein C (APC) or in binding C4BP. Additionally, protein S has a peptide between the Gla domain and the EGF-like domain, that is cleaved by thrombin. The Gla and EGF-like domains stay connected after the cleavage by a disulfide bond. However, protein S loses its function as an APC cofactor following either this cleavage or binding C4BP.
- Stenflo J (1999). “Contributions of Gla and EGF-like domains to the function of vitamin K-dependent coagulation factors”. Critical Reviews in Eukaryotic Gene Expression. 9 (1): 59–88. doi:10.1615/CritRevEukaryotGeneExpr.v9.i1.50. PMID 10200912.
- Rosner W (Dec 1991). “Plasma steroid-binding proteins”. Endocrinology and Metabolism Clinics of North America. 20 (4): 697–720. doi:10.1016/S0889-8529(18)30240-8. PMID 1778174.
- Dahlbäck B, Lundwall A, Stenflo J (Jun 1986). “Primary structure of bovine vitamin K-dependent protein S”. Proceedings of the National Academy of Sciences. 83 (12): 4199–203. Bibcode:1986PNAS…83.4199D. doi:10.1073/pnas.83.12.4199. PMC 323699. PMID 2940598
- The best characterized function of Protein S is its role in the anti coagulation pathway, where it functions as a cofactor to Protein C in the inactivation of Factors Va and VIIIa. Only the free form has cofactor activity. Protein S binds to negatively charged phospholipids via the carboxylated Gla domain. This property allows Protein S to facilitate the removal of cells that are undergoing apoptosis, a form of structured cell death used by the body to remove unwanted or damaged cells. In healthy cells, an ATP (adenosine triphosphate)-dependent enzyme removes negatively charged phospholipids such as phosphatidyl serine from the outer leaflet of the cell membrane. An apoptotic cell (that is, one undergoing apoptosis) no longer actively manages the distribution of phospholipids in its outer membrane and hence begins to display negatively charged phospholipids on its exterior surface. These negatively charged phospholipids are recognized by phagocytes such as macrophages. Protein S binds to the negatively charged phospholipids and functions as a bridge between the apoptotic cell and the phagocyte. This bridging expedites phagocytosis and allows the cell to be removed without giving rise to inflammation or other signs of tissue damage.
- Protein S does not bind to the nascent complement complex C5,6,7 to prevents it from inserting into a membrane. This is a different complement protein S AKA vitronectin made by the VTN gene, not to be confused with the coagulation protein S made by the PROS gene which this wiki page concerns.
- Vitronectin (VTN or VN) is a glycoprotein of the hemopexin family which is synthesized and excreted by the liver, and abundantly found in serum, the extracellular matrix and bone. In humans it is encoded by the VTN gene.
- Boron, Walter F. and Boulpaep, Emile L. “Medical Physiology”. Saunders, 2012, p.1097.
- “Entrez Gene: M Vitronectin”.
- Jenne D, Stanley KK (Oct 1987). “Nucleotide sequence and organization of the human S-protein gene: repeating peptide motifs in the “pexin” family and a model for their evolution”. Biochemistry. 26 (21): 6735–42. doi:10.1021/bi00395a024. PMID 2447940.
- Vitronectin binds to integrin alpha-V beta-3 and thus promotes cell adhesion and spreading. It also inhibits the membrane-damaging effect of the terminal cytolytic complement pathway and binds to several serpins (serine protease inhibitors). It is a secreted protein and exists in either a single chain form or a clipped, two chain form held together by a disulfide bond. Vitronectin has been speculated to be involved in hemostasis and tumor malignancy.
- Preissner KT, Seiffert D (Jan 1998). “Role of vitronectin and its receptors in haemostasis and vascular remodeling”. Thrombosis Research. 89 (1): 1–21. doi:10.1016/S0049-3848(97)00298-3. PMID 9610756.
- Felding-Habermann B, Cheresh DA (Oct 1993). “Vitronectin and its receptors”. Current Opinion in Cell Biology. 5 (5): 864–8. doi:10.1016/0955-0674(93)90036-P. PMID 7694604.
- Hurt, Elaine M.; Chan, King; Serrat, Maria Ana Duhagon; Thomas, Suneetha B.; Veenstra, Timothy D.; Farrar, William L. (2009). “Identification of Vitronectin as an Extrinsic Inducer of Cancer Stem Cell Differentiation and Tumor Formation”. Stem Cells. 28 (3): 390–8. doi:10.1002/stem.271. PMC 3448441. PMID 19998373.
- Vitronectin is a 54 kDa glycoprotein, consisting of 478 amino acid residues. About one-third of the protein’s molecular mass is composed of carbohydrates. On occasion, the protein is cleaved after arginine 379, to produce two-chain vitronectin, where the two parts are linked by a disulfide bond. No high-resolution structure has been determined experimentally yet, except for the N-terminal domain.
- The protein consists of three domains:
- The N-terminal Somatomedin B domain (1-39)
- A central domains with hemopexin homology (131-342)
- A C-terminal domain (residues 347-459) also with hemopexin homology.
- Several structures has been reported for the Somatomedin B domain. The protein was initially crystallized in complex with one of its physiological binding partners: the Plasminogen activator inhibitor-1 (PAI-1) and the structure solved for this complex. Subsequently two groups reported NMR structures of the domain.
- Zhou A, Huntington JA, Pannu NS, Carrell RW, Read RJ (Jul 2003). “How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration”. Nature Structural Biology. 10 (7): 541–4. doi:10.1038/nsb943. PMID 12808446. S2CID 26086796.
- Kamikubo Y, De Guzman R, Kroon G, Curriden S, Neels JG, Churchill MJ, Dawson P, Ołdziej S, Jagielska A, Scheraga HA, Loskutoff DJ, Dyson HJ (Jun 2004). “Disulfide bonding arrangements in active forms of the somatomedin B domain of human vitronectin”. Biochemistry. 43 (21): 6519–34. doi:10.1021/bi049647c. PMID 15157085.
- Mayasundari A, Whittemore NA, Serpersu EH, Peterson CB (Jul 2004). “The solution structure of the N-terminal domain of human vitronectin: proximal sites that regulate fibrinolysis and cell migration”. The Journal of Biological Chemistry. 279 (28): 29359–66. doi:10.1074/jbc.M401279200. PMID 15123712.
- The somatomedin B domain is a close-knit disulfide knot, with 4 disulfide bonds within 35 residues. Different disulfide configurations had been reported for this domain but this ambiguity has been resolved by the crystal structure. Homology models have been built for the central and C-terminal domains.
- Kamikubo Y, Okumura Y, Loskutoff DJ (Jul 2002). “Identification of the disulfide bonds in the recombinant somatomedin B domain of human vitronectin”. The Journal of Biological Chemistry. 277 (30): 27109–19. doi:10.1074/jbc.M200354200. PMID 12019263.
- Horn NA, Hurst GB, Mayasundari A, Whittemore NA, Serpersu EH, Peterson CB (Aug 2004). “Assignment of the four disulfides in the N-terminal somatomedin B domain of native vitronectin isolated from human plasma”. The Journal of Biological Chemistry. 279 (34): 35867–78. doi:10.1074/jbc.M405716200. PMID 15173163.
- Xu D, Baburaj K, Peterson CB, Xu Y (Aug 2001). “Model for the three-dimensional structure of vitronectin: predictions for the multi-domain protein from threading and docking”. Proteins. 44 (3): 312–20. doi:10.1002/prot.1096. PMID 11455604. S2CID 24765480.
- The somatomedin B domain of vitronectin binds to plasminogen activator inhibitor-1 (PAI-1), and stabilizes it. Thus vitronectin serves to regulate proteolysis initiated by plasminogen activation. In addition, vitronectin is a component of platelets and is, thus, involved in hemostasis. Vitronectin contains an RGD (45-47) sequence, which is a binding site for membrane-bound integrins, e.g., the vitronectin receptor, which serve to anchor cells to the extracellular matrix. The Somatomedin B domain interacts with the urokinase receptor, and this interaction has been implicated in cell migration and signal transduction. High plasma levels of both PAI-1 and the urokinase receptor have been shown to correlate with a negative prognosis for cancer patients. Cell adhesion and migration are directly involved in cancer metastasis, which provides a probable mechanistic explanation for this observation.
- αVβ3 is a type of integrin that is a receptor for vitronectin. It consists of two components, integrin alpha V and integrin beta 3 (CD61), and is expressed by platelets. Furthermore, it is a receptor for phagocytosis on macrophages or dendritic cells. Integrin αVβ3 is a potential drug target because abnormal expression of v3 is linked to the development and progression of various diseases. Its role in angiogenesis, in cancer and other diseases, is linked to the blood supply for problematic overgrowths. Inhibitors like etaracizumab may be used as antiangiogenics. One novel protein (ProAgio) has been designed to bind at an unusual site, and then induces apoptosis by recruiting caspase 8. It is designed by mutating domain 1 of CD2 (D1-CD2), which naturally binds weakly to the receptor. Fibronectin domain 10 contains the RGD motif that αVβ3 recognizes. A high-affinity, pure antagonist mutant has been discovered for this protein.
- Hermann P, Armant M, Brown E, Rubio M, Ishihara H, Ulrich D, Caspary RG, Lindberg FP, Armitage R, Maliszewski C, Delespesse G, Sarfati M (February 1999). “The vitronectin receptor and its associated CD47 molecule mediates proinflammatory cytokine synthesis in human monocytes by interaction with soluble CD23”. The Journal of Cell Biology. 144 (4): 767–75. doi:10.1083/jcb.144.4.767. PMC 2132927. PMID 10037797.
- Yamaguchi H, Takagi J, Miyamae T, Yokota S, Fujimoto T, Nakamura S, Ohshima S, Naka T, Nagata S (May 2008). “Milk fat globule EGF factor 8 in the serum of human patients of systemic lupus erythematosus”. Journal of Leukocyte Biology. 83 (5): 1300–7. doi:10.1189/jlb.1107730. PMID 18303131.
- Novel Protein Agent Targets Cancer and Host of Other Diseases. June 2016
- Santulli G, Basilicata MF, De Simone M, Del Giudice C, Anastasio A, Sorriento D, Saviano M, Del Gatto A, Trimarco B, Pedone C, Zaccaro L, Iaccarino G (January 2011). “Evaluation of the anti-angiogenic properties of the new selective αVβ3 integrin antagonist RGDechiHCit”. Journal of Translational Medicine. 9 (1): 7. doi:10.1186/1479-5876-9-7. PMC 3027097. PMID 21232121.
- Turaga, Ravi Chakra; Yin, Lu; Yang, Jenny J.; Lee, Hsiauwei; Ivanov, Ivaylo; Yan, Chunli; Yang, Hua; Grossniklaus, Hans E.; Wang, Siming; Ma, Cheng; Sun, Li; Liu, Zhi-Ren (31 May 2016). “Rational design of a protein that binds integrin αvβ3 outside the ligand binding site”. Nature Communications. 7 (1): 11675. Bibcode:2016NatCo…711675T. doi:10.1038/ncomms11675. PMC 4895024. PMID 27241473.
- Van Agthoven JF, Xiong JP, Alonso JL, Rui X, Adair BD, Goodman SL, Arnaout MA (April 2014). “Structural basis for pure antagonism of integrin αVβ3 by a high-affinity form of fibronectin”. Nature Structural & Molecular Biology. 21 (4): 383–8. doi:10.1038/nsmb.2797. PMC 4012256. PMID 24658351.
- Vitaxin (MEDI-523) is a humanized monoclonal antibody against the vascular integrin alpha-v beta-3. It is shown to be a promising angiogenesis inhibitor used in the treatment of some forms of cancer. Vitaxin was in 2002 being studied for rheumatoid arthritis. It is the developmental precursor of Etaracizumab (MEDI-522). Both are derived from the mouse antibody LM609.
- Cherrington JM, Strawn LM, Shawver LK (2000). Vande Woude GF, Klein G (eds.). “New paradigms for the treatment of cancer: the role of anti-angiogenesis agents”. Advances in Cancer Research. Academic Press. 79: 1–38 (27). doi:10.1016/s0065-230x(00)79001-4. ISBN 978-0-12-006679-7. PMID 10818676.
- Wilder RL (November 2002). “Integrin alpha V beta 3 as a target for treatment of rheumatoid arthritis and related rheumatic diseases”. Annals of the Rheumatic Diseases. 61 (Suppl 2): ii96–ii99. doi:10.1136/ard.61.suppl_2.ii96. PMC 1766704. PMID 12379637.
- McNeel DG, Eickhoff J, Lee FT, King DM, Alberti D, Thomas JP, et al. (November 2005). “Phase I trial of a monoclonal antibody specific for alphavbeta3 integrin (MEDI-522) in patients with advanced malignancies, including an assessment of effect on tumor perfusion”. Clinical Cancer Research. 11 (21): 7851–7860. doi:10.1158/1078-0432.CCR-05-0262. PMID 16278408. S2CID 33227412.
- Vitaxin (MEDI-523) is a humanized monoclonal antibody against the vascular integrin alpha-v beta-3. It is shown to be a promising angiogenesis inhibitor used in the treatment of some forms of cancer. Vitaxin was in 2002 being studied for rheumatoid arthritis. It is the developmental precursor of Etaracizumab (MEDI-522). Both are derived from the mouse antibody LM609.
- The Urokinase receptor, also known as urokinase plasminogen activator surface receptor (uPAR) or CD87 (Cluster of Differentiation 87), is a protein encoded in humans by the PLAUR gene. It is a multidomain glycoprotein tethered to the cell membrane with a glycosylphosphotidylinositol (GPI) anchor. uPAR was originally identified as a saturable binding site for urokinase (also known as uPA) on the cell surface. uPAR consists of three tandem LU domains, which are protein domains of the three-finger protein family. The structure of uPAR has been solved by X-ray crystallography in complex with a peptide antagonist and with its native ligand, urokinase. All three three-finger domains are necessary for high affinity binding of the primary ligand, urokinase. In addition, uPAR also interacts with several other proteins, including vitronectin, the uPAR associated protein (uPARAP) and the integrin family of membrane proteins. It has been possible to express uPAR recombinantly in CHO-cells and S2 cells from Drosophila melanogaster. 4 out of 5 of the possible glycosylation sites are used in vivo giving the protein a molecular weight of 50–60 kDA.
- Kessler, Pascal; Marchot, Pascale; Silva, Marcela; Servent, Denis (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142: 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- Llinas P, Le Du MH, Gårdsvoll H, Danø K, Ploug M, Gilquin B, Stura EA, Ménez A (May 2005). “Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide”. The EMBO Journal. 24 (9): 1655–63. doi:10.1038/sj.emboj.7600635. PMC 1142576. PMID 15861141.
- Huai Q, Mazar AP, Kuo A, Parry GC, Shaw DE, Callahan J, Li Y, Yuan C, Bian C, Chen L, Furie B, Furie BC, Cines DB, Huang M (February 2006). “Structure of human urokinase plasminogen activator in complex with its receptor”. Science. 311 (5761): 656–9. Bibcode:2006Sci…311..656H. doi:10.1126/science.1121143. PMID 16456079. S2CID 39521660.
- uPAR is a part of the plasminogen activation system, which in the healthy body is involved in tissue reorganization events such as mammary gland involution and wound healing. In order to be able to reorganize tissue, the old tissue must be able to be degraded. An important mechanism in this degradation is the proteolysis cascade initiated by the plasminogen activation system. uPAR binds urokinase and thus restricts plasminogen activation to the immediate vicinity of the cell membrane. When urokinase is bound to the receptor, there is cleavage between the GPI-anchor and the uPAR, releasing a soluble form of the protein known as suPAR.
- ViroGates. “What is suPAR”. suPARnostic® by ViroGates. Retrieved 2021-09-27.
- Thunø M, Macho B, Eugen-Olsen J (2009). “suPAR: the molecular crystal ball”. Disease Markers. 27 (3): 157–72. doi:10.1155/2009/504294. PMC 3835059. PMID 19893210.
- Soluble urokinase plasminogen activator receptor (suPAR) has been found to be a biomarker of inflammation. Elevated suPAR is seen in chronic obstructive pulmonary disease, asthma, liver failure, heart failure, cardiovascular disease, and rheumatoid arthritis.Smokers have significantly higher suPAR compared to non-smokers.
- Desmedt S, Desmedt V, Delanghe JR, Speeckaert R, Speeckaert MM (2017). “The Intriguing Role of Soluble Urokinase Receptor in Inflammatory Diseases”. Critical Reviews in Clinical Laboratory Sciences. 54 (2): 117–133. doi:10.1080/10408363.2016.1269310. PMID 28084848. S2CID 32624995.
- Urokinase receptors have been found to be highly expressed on senescent cells, leading researchers to use chimeric antigen receptor T cells to eliminate senescent cells in mice.
- Wagner V, Gil J (2020). “T Cells Engineered to Target Senescence”. Nature. 583 (7814): 37–38. Bibcode:2020Natur.583…37W. doi:10.1038/d41586-020-01759-x. hdl:10044/1/80980. PMID 32601490.
- ^ Amor C, Feucht J, Lowe SW (2020). “Senolytic CAR T cells reverse senescence-associated pathologies”. Nature. 583 (7814): 127–132. doi:10.1038/s41586-020-2403-9. PMC 7583560. PMID 32555459.
- The components of the plasminogen activation system have been found to be highly expressed in many malignant tumors, indicating that tumors are able to hijack the system, and use it in metastasis. Thus inhibitors of the various components of the plasminogen activation system have been sought as possible anticancer drugs.
- Josip Madunić (2018). “The Urokinase Plasminogen Activator System in Human Cancers: An Overview of Its Prognostic and Predictive Role”. Thrombosis and Haemostasis. 118 (12): 2020–2036. doi:10.1055/s-0038-1675399. PMID 30419600.
- uPAR has been involved in various other non-proteolytic processes related to cancer, such as cell migration, cell cycle regulation, and cell adhesion. Urokinase receptor has been shown to interact with LRP1.
- Czekay RP, Kuemmel TA, Orlando RA, Farquhar MG (May 2001). “Direct binding of occupied urokinase receptor (uPAR) to LDL receptor-related protein is required for endocytosis of uPAR and regulation of cell surface urokinase activity”. Molecular Biology of the Cell. 12 (5): 1467–79. doi:10.1091/mbc.12.5.1467. PMC 34598. PMID 11359936.
- Vitronectin (VTN or VN) is a glycoprotein of the hemopexin family which is synthesized and excreted by the liver, and abundantly found in serum, the extracellular matrix and bone. In humans it is encoded by the VTN gene.
- Mutations in the PROS1 gene can lead to Protein S deficiency which is a rare blood disorder which can lead to an increased risk of thrombosis. See also Hemostasis
- Beauchamp NJ, Dykes AC, Parikh N, Campbell Tait R, Daly ME (June 2004). “The prevalence of, and molecular defects underlying, inherited protein S deficiency in the general population”. Br. J. Haematol. 125 (5): 647–54. doi:10.1111/j.1365-2141.2004.04961.x. PMID 15147381. S2CID 705661.
- García de Frutos P, Fuentes-Prior P, Hurtado B, Sala N (September 2007). “Molecular basis of protein S deficiency”. Thromb. Haemost. 98 (3): 543–56. doi:10.1160/th07-03-0199. PMID 17849042. S2CID 17274778.
- Protein S (also known as PROS) is a vitamin K-dependent plasma glycoprotein synthesized in the liver. In the circulation, Protein S exists in two forms: a free form and a complex form bound to complement protein C4b-binding protein (C4BP). In humans, protein S is encoded by the PROS1 gene. Protein S plays a role in coagulation. Protein S is named for Seattle, Washington, where it was originally discovered and purified by Earl Davie‘s group in 1977.
- Glycoprotein Ib (GPIb), also known as CD42, is a component of the GPIb-V-IX complex on platelets. The GPIb-V-IX complex binds von Willebrand factor, allowing platelet adhesion and platelet plug formation at sites of vascular injury. Glycoprotein Ibα (GPIbα) is the major ligand-binding subunit of the GPIb-V-IX complex. GPIbα is heavily glycosylated. It is deficient in the Bernard–Soulier syndrome. A gain-of-function mutation causes platelet-type von Willebrand disease. Autoantibodies against Ib/IX can be produced in immune thrombocytopenic purpura. Components include GP1BA and GP1BB. It complexes with Glycoprotein IX.
- This binding occurs under all circumstances, but is most efficient under high shear stress (i.e., rapid blood flow in narrow blood vessels, see below).
- VWF binds to other platelet receptors when they are activated, e.g., by thrombin (i.e., when coagulation has been stimulated).
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID 9759493.
VWF plays a major role in blood coagulation. Therefore, VWF deficiency or dysfunction (von Willebrand disease) leads to a bleeding tendency, which is most apparent in tissues having high blood flow shear in narrow vessels. From studies it appears that VWF uncoils under these circumstances, decelerating passing platelets. Recent research also suggests that von Willebrand Factor is involved in the formation of blood vessels themselves, which would explain why some people with von Willebrand disease develop vascular malformations (predominantly in the digestive tract) that can bleed excessively.
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID 9759493.
- Randi AM, Laffan MA (January 2017). “Von Willebrand factor and angiogenesis: basic and applied issues”. Journal of Thrombosis and Haemostasis. 15 (1): 13–20. doi:10.1111/jth.13551. hdl:10044/1/42796. PMID 27778439. S2CID 3490036.
Catabolism
The biological breakdown (catabolism) of VWF is largely mediated by the enzyme ADAMTS13 (acronym of “a disintegrin-like and metalloprotease with thrombospondin type 1 motif no. 13“). It is a metalloproteinase that cleaves VWF between tyrosine at position 842 and methionine at position 843 (or 1605–1606 of the gene) in the A2 domain. This breaks down the multimers into smaller units, which are degraded by other peptidases.
- Levy GG, Motto DG, Ginsburg D (July 2005). “ADAMTS13 turns 3”. Blood. 106 (1): 11–17. doi:10.1182/blood-2004-10-4097. PMID 15774620. S2CID 25645477.
The half-life of vWF in human plasma is around 16 hours; glycosylation variation on vWF molecules from different individuals result in a larger range of 4.2 to 26 hours. Liver cells as well as macrophages take up vWF for clearance via ASGPRs and LRP1. SIGLEC5 and CLEC4M also recognize vWF.
- Lenting PJ, Christophe OD, Denis CV (March 2015). “von Willebrand factor biosynthesis, secretion, and clearance: connecting the far ends”. Blood. 125 (13): 2019–2028. doi:10.1182/blood-2014-06-528406. PMID 25712991. S2CID 27785232.
Role in disease
Main article: von Willebrand disease
Hereditary or acquired defects of VWF lead to von Willebrand disease (vWD), a bleeding diathesis of the skin and mucous membranes, causing nosebleeds, menorrhagia, and gastrointestinal bleeding. The point at which the mutation occurs determines the severity of the bleeding diathesis. There are three types (I, II and III), and type II is further divided in several subtypes. Treatment depends on the nature of the abnormality and the severity of the symptoms. Most cases of vWD are hereditary, but abnormalities of VWF may be acquired; aortic valve stenosis, for instance, has been linked to vWD type IIA, causing gastrointestinal bleeding – an association known as Heyde’s syndrome.
- Sadler JE, Budde U, Eikenboom JC, Favaloro EJ, Hill FG, Holmberg L, et al. (October 2006). “Update on the pathophysiology and classification of von Willebrand disease: a report of the Subcommittee on von Willebrand Factor”. Journal of Thrombosis and Haemostasis. 4 (10): 2103–2114. doi:10.1111/j.1538-7836.2006.02146.x. PMID 16889557. S2CID 23875096.
- Vincentelli A, Susen S, Le Tourneau T, Six I, Fabre O, Juthier F, et al. (July 2003). “Acquired von Willebrand syndrome in aortic stenosis”. The New England Journal of Medicine. 349 (4): 343–349. doi:10.1056/NEJMoa022831. PMID 12878741.
In thrombotic thrombocytopenic purpura (TTP) and hemolytic–uremic syndrome (HUS), ADAMTS13 either is deficient or has been inhibited by antibodies directed at the enzyme. This leads to decreased breakdown of the ultra-large multimers of VWF and microangiopathic hemolytic anemia with deposition of fibrin and platelets in small vessels, and capillary necrosis. In TTP, the organ most obviously affected is the brain; in HUS, the kidney.
- Moake JL (January 2004). “von Willebrand factor, ADAMTS-13, and thrombotic thrombocytopenic purpura”. Seminars in Hematology. 41 (1): 4–14. doi:10.1053/j.seminhematol.2003.10.003. PMID 14727254.
Higher levels of VWF are more common among people that have had ischemic stroke (from blood-clotting) for the first time. Occurrence is not affected by ADAMTS13, and the only significant genetic factor is the person’s blood group. High plasma VWF levels were found to be an independent predictor of major bleeding in anticoagulated atrial fibrillation patients.
- Denorme F, De Meyer SF (September 2016). “The VWF-GPIb axis in ischaemic stroke: lessons from animal models”. Thrombosis and Haemostasis. 116 (4): 597–604. doi:10.1160/TH16-01-0036. PMID 27029413. S2CID 4964177.
- Roldán V, Marín F, Muiña B, Torregrosa JM, Hernández-Romero D, Valdés M, et al. (June 2011). “Plasma von Willebrand factor levels are an independent risk factor for adverse events including mortality and major bleeding in anticoagulated atrial fibrillation patients”. Journal of the American College of Cardiology. 57 (25): 2496–2504. doi:10.1016/j.jacc.2010.12.033. PMID 21497043.
History
See also: Erik Adolf von Willebrand § Von Willebrand disease
VWF is named after Erik Adolf von Willebrand, a Finnish physician who in 1926 first described a hereditary bleeding disorder in families from Åland. Although von Willebrand did not identify the definite cause, he distinguished von Willebrand disease (vWD) from hemophilia and other forms of bleeding diathesis.
- von Willebrand EA (1926). “Hereditär pseudohemofili” [Hereditary pseudo haemophilia]. Fin Läkaresällsk Handl (in Swedish). 68: 87–112. Reproduced in Von Willebrand EA (May 1999). “Hereditary pseudohaemophilia”. Haemophilia. 5 (3): 223–31, discussion 222. doi:10.1046/j.1365-2516.1999.00302.x. PMID 10444294. S2CID 221750622.
In the 1950s, vWD was shown to be caused by a plasma factor deficiency (instead of being caused by platelet disorders), and, in the 1970s, the VWF protein was purified. Harvey J. Weiss and coworkers developed a quantitative assay for VWF function that remains a mainstay of laboratory evaluation for VWD to this day.
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID 9759493.
- Weiss HJ, Hoyer IW (December 1973). “Von Willebrand factor: dissociation from antihemophilic factor procoagulant activity”. Science. 182 (4117): 1149–1151. Bibcode:1973Sci…182.1149W. doi:10.1126/science.182.4117.1149. PMID 4127287. S2CID 41340436.
- Weiss HJ, Rogers J, Brand H (November 1973). “Defective ristocetin-induced platelet aggregation in von Willebrand’s disease and its correction by factor VIII”. The Journal of Clinical Investigation. 52 (11): 2697–2707. doi:10.1172/JCI107464. PMC 302536. PMID 4201262.
Interactions
Von Willebrand Factor has been shown to interact with Collagen, type I, alpha 1.
- Pareti FI, Fujimura Y, Dent JA, Holland LZ, Zimmerman TS, Ruggeri ZM (November 1986). “Isolation and characterization of a collagen binding domain in human von Willebrand factor”. The Journal of Biological Chemistry. 261 (32): 15310–15315. doi:10.1016/S0021-9258(18)66869-3. PMID 3490481.
Collagen, type I, alpha 1, also known as alpha-1 type I collagen, is a protein that in humans is encoded by the COL1A1 gene. COL1A1 encodes the major component of type I collagen, the fibrillar collagen found in most connective tissues, including cartilage.
Function
Collagen is a protein that strengthens and supports many tissues in the body, including cartilage, bone, tendon, skin and the white part of the eye (sclera). The COL1A1 gene produces a component of type I collagen, called the pro-alpha1(I) chain. This chain combines with another pro-alpha1(I) chain and also with a pro-alpha2(I) chain (produced by the COL1A2 gene) to make a molecule of type I procollagen. These triple-stranded, rope-like procollagen molecules must be processed by enzymes outside the cell. Once these molecules are processed, they arrange themselves into long, thin fibrils that cross-link to one another in the spaces around cells. The cross-links result in the formation of very strong mature type I collagen fibers. Collagenous function includes rigidity and elasticity.
Gene
The COL1A1 gene is located on the long (q) arm of chromosome 17 between positions 21.3 and 22.1, from base pair 50183289 to base pair 50201632.
Clinical significance
Mutations in the COL1A1 gene are associated with the following conditions:
- Ehlers–Danlos syndrome, vascular type: In rare cases, specific heterozygous arginine-to-cysteine substitution mutations in COL1A1 that are also associated with vascular fragility and mimic COL3A1-vEDS
- Ehlers–Danlos syndrome, arthrochalasia type: It is caused by mutations in the COL1A1 gene. The mutations in the COL1A1 gene that cause this disorder instruct the cell to leave out a part of the pro-alpha1(I) chain that contains a segment used to attach one molecule to another. When this part of the protein is missing, the structure of type I collagen is compromised. Tissues that are rich in type I collagen, such as the skin, bones, and tendons, are affected by this change. Ehlers–Danlos type IV is most attributed to abnormalities in the reticular fibers (collagen Type III).
- Ehlers–Danlos syndrome, classical type: In rare cases, a mutation in the COL1A1 gene has been shown to cause the classical type of Ehlers–Danlos syndrome. This mutation substitutes the amino acid cysteine for the amino acid arginine at position 134 in the protein made by the gene. (The mutation can also be written as Arg134Cys.) The altered protein interacts abnormally with other collagen-building proteins, disrupting the structure of type I collagen fibrils and trapping collagen in the cell. Researchers believe that these changes in collagen cause the signs and symptoms of the disorder. Ehlers–Danlos type IV is most attributed to abnormalities in the reticular fibers (collagen Type III). Without the hydroxylation of lysine, by the enzyme lysyl hydroxylase, the final collagen structure cannot form.
- Osteogenesis imperfecta, type I: Osteogenesis imperfecta is the most common disorder caused by mutations in this gene. Mutations that inactivate one of the two copies of the COL1A1 gene cause osteogenesis imperfecta type I. The mutated copy of the gene does not produce any pro-alpha1(I) collagen chains. Because only one copy of the gene is directing the cell to make pro-alpha1(I) chains, cells from people with this disorder make only half of the normal amount of type I collagen, which results in bone fragility and other symptoms.
- Osteogenesis imperfecta, type II: Many different types of mutations in the COL1A1 gene can cause osteogenesis imperfecta type II. These mutations range from missing pieces of the COL1A1 gene to amino acid substitutions, in which the amino acid glycine is replaced by another amino acid in the protein strand. Sometimes one end of the gene (called the C-terminus) is altered, which interferes with the association of the protein strands. All of these changes prevent the normal production of mature type I collagen, which results in this severe condition, type II osteogenesis imperfecta.
- Osteogenesis imperfecta, type III: Mutations in the COL1A1 gene may result in the production of a protein that is missing segments, making it unusable for collagen production. Other mutations cause the amino acid glycine to be replaced by a different amino acid in the pro-alpha1(I) chain, which inhibits the essential interaction between protein chains. Type I collagen production is inhibited by the inability of the altered procollagen strands to associate and form the triple-stranded, ropelike structure of mature collagen. These alterations negatively affect tissues that are rich in type I collagen, such as the skin, bones, teeth, and tendons, leading to the signs and symptoms of type III osteogenesis imperfecta.
- Osteogenesis imperfecta, type IV: Several different types of mutations in the COL1A1 gene cause osteogenesis imperfecta type IV. These mutations may involve missing pieces of the COL1A1 gene or changes in base pairs (the building blocks of DNA). These gene alterations result in a protein that is missing segments or has amino acid substitutions; specifically, the amino acid glycine is replaced by another amino acid. All of these changes interfere with the formation of the mature triple-stranded collagen molecule and prevent the production of mature type I collagen, which results in type IV osteogenesis imperfecta.
- Osteoporosis: Osteoporosis is a condition that makes bones progressively more brittle and prone to fracturing. A particular variation (polymorphism) in the COL1A1 gene appears to increase the risk of developing osteoporosis. A specific variation at Sp1 binding site is shown to be associated with increased risk of low bone mass and vertebral fracture, because of the changes the COL1A1 protein produced from one copy of the gene. Several studies have shown that women with this particular genetic variation at Sp1 site are more likely to have signs of osteoporosis than are women without the variation.
- Predisposition to hernias.
- Sezer S, Şimşek N, Celik HT, Erden G, Ozturk G, Düzgün AP, Çoşkun F, Demircan K (2014). “Association of collagen type I alpha 1 gene polymorphism with inguinal hernia – PubMed”. Hernia: The Journal of Hernias and Abdominal Wall Surgery. 18 (4): 507–12. doi:10.1007/s10029-013-1147-y. PMID 23925543. S2CID 22999363.
- Predisposition to degenerative disc disease, and disc herniation.
- Kawaguchi Y (2018). “Genetic background of degenerative disc disease in the lumbar spine”. Spine Surgery and Related Research. 2 (2): 98–112. doi:10.22603/ssrr.2017-0007. PMC 6698496. PMID 31440655
External links
- GeneReviews/NCBI/NIH/UW entry on Osteogenesis Imperfecta
- Online Mendelian Inheritance in Man (OMIM): 120150
- EntrezGene 1277
- COL1A1 GeneCard
- Database of human type I and type III collagen mutations
- Overview of all the structural information available in the PDB for UniProt: P02452 (Collagen alpha-1(I) chain) at the PDBe-KB.
Recently, It has been reported that the cooperation and interactions within the von Willebrand Factors enhances the adsorption probability in the primary haemostasis. Such cooperation is proven by calculating the adsorption probability of flowing VWF once it crosses another adsorbed one. Such cooperation is held within a wide range of shear rates.
- Heidari M, Mehrbod M, Ejtehadi MR, Mofrad MR (August 2015). “Cooperation within von Willebrand factors enhances adsorption mechanism”. Journal of the Royal Society, Interface. 12 (109): 20150334. doi:10.1098/rsif.2015.0334. PMC 4535404. PMID 26179989.
See also
References
- GRCh38: Ensembl release 89: ENSG00000110799 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000001930 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- Sadler JE (1998). “Biochemistry and genetics of von Willebrand factor”. Annual Review of Biochemistry. 67: 395–424. doi:10.1146/annurev.biochem.67.1.395. PMID 9759493.
- Shahidi M (2017). “Thrombosis and von Willebrand Factor”. Thrombosis and Embolism: From Research to Clinical Practice. Advances in Experimental Medicine and Biology. Vol. 906. pp. 285–306. doi:10.1007/5584_2016_122. ISBN 978-3-319-22107-6. PMID 27628010.
- Zhou YF, Eng ET, Zhu J, Lu C, Walz T, Springer TA (July 2012). “Sequence and structure relationships within von Willebrand factor”. Blood. 120 (2): 449–458. doi:10.1182/blood-2012-01-405134. PMC 3398765. PMID 22490677.
- Jakobi AJ, Mashaghi A, Tans SJ, Huizinga EG (July 2011). “Calcium modulates force sensing by the von Willebrand factor A2 domain”. Nature Communications. 2: 385. Bibcode:2011NatCo…2..385J. doi:10.1038/ncomms1385. PMC 3144584. PMID 21750539.
- Luken BM, Winn LY, Emsley J, Lane DA, Crawley JT (June 2010). “The importance of vicinal cysteines, C1669 and C1670, for von Willebrand factor A2 domain function”. Blood. 115 (23): 4910–4913. doi:10.1182/blood-2009-12-257949. PMC 2890177. PMID 20354169.
- Lenting PJ, Christophe OD, Denis CV (March 2015). “von Willebrand factor biosynthesis, secretion, and clearance: connecting the far ends”. Blood. 125 (13): 2019–2028. doi:10.1182/blood-2014-06-528406. PMID 25712991. S2CID 27785232.
- Randi AM, Laffan MA (January 2017). “Von Willebrand factor and angiogenesis: basic and applied issues”. Journal of Thrombosis and Haemostasis. 15 (1): 13–20. doi:10.1111/jth.13551. hdl:10044/1/42796. PMID 27778439. S2CID 3490036.
- Levy GG, Motto DG, Ginsburg D (July 2005). “ADAMTS13 turns 3”. Blood. 106 (1): 11–17. doi:10.1182/blood-2004-10-4097. PMID 15774620. S2CID 25645477.
- Sadler JE, Budde U, Eikenboom JC, Favaloro EJ, Hill FG, Holmberg L, et al. (October 2006). “Update on the pathophysiology and classification of von Willebrand disease: a report of the Subcommittee on von Willebrand Factor”. Journal of Thrombosis and Haemostasis. 4 (10): 2103–2114. doi:10.1111/j.1538-7836.2006.02146.x. PMID 16889557. S2CID 23875096.
- Vincentelli A, Susen S, Le Tourneau T, Six I, Fabre O, Juthier F, et al. (July 2003). “Acquired von Willebrand syndrome in aortic stenosis”. The New England Journal of Medicine. 349 (4): 343–349. doi:10.1056/NEJMoa022831. PMID 12878741.
- Moake JL (January 2004). “von Willebrand factor, ADAMTS-13, and thrombotic thrombocytopenic purpura”. Seminars in Hematology. 41 (1): 4–14. doi:10.1053/j.seminhematol.2003.10.003. PMID 14727254.
- Denorme F, De Meyer SF (September 2016). “The VWF-GPIb axis in ischaemic stroke: lessons from animal models”. Thrombosis and Haemostasis. 116 (4): 597–604. doi:10.1160/TH16-01-0036. PMID 27029413. S2CID 4964177.
- Roldán V, Marín F, Muiña B, Torregrosa JM, Hernández-Romero D, Valdés M, et al. (June 2011). “Plasma von Willebrand factor levels are an independent risk factor for adverse events including mortality and major bleeding in anticoagulated atrial fibrillation patients”. Journal of the American College of Cardiology. 57 (25): 2496–2504. doi:10.1016/j.jacc.2010.12.033. PMID 21497043.
- von Willebrand EA (1926). “Hereditär pseudohemofili” [Hereditary pseudo haemophilia]. Fin Läkaresällsk Handl (in Swedish). 68: 87–112. Reproduced in Von Willebrand EA (May 1999). “Hereditary pseudohaemophilia”. Haemophilia. 5 (3): 223–31, discussion 222. doi:10.1046/j.1365-2516.1999.00302.x. PMID 10444294. S2CID 221750622.
- Weiss HJ, Hoyer IW (December 1973). “Von Willebrand factor: dissociation from antihemophilic factor procoagulant activity”. Science. 182 (4117): 1149–1151. Bibcode:1973Sci…182.1149W. doi:10.1126/science.182.4117.1149. PMID 4127287. S2CID 41340436.
- Weiss HJ, Rogers J, Brand H (November 1973). “Defective ristocetin-induced platelet aggregation in von Willebrand’s disease and its correction by factor VIII”. The Journal of Clinical Investigation. 52 (11): 2697–2707. doi:10.1172/JCI107464. PMC 302536. PMID 4201262.
- Pareti FI, Fujimura Y, Dent JA, Holland LZ, Zimmerman TS, Ruggeri ZM (November 1986). “Isolation and characterization of a collagen binding domain in human von Willebrand factor”. The Journal of Biological Chemistry. 261 (32): 15310–15315. doi:10.1016/S0021-9258(18)66869-3. PMID 3490481.
- Heidari M, Mehrbod M, Ejtehadi MR, Mofrad MR (August 2015). “Cooperation within von Willebrand factors enhances adsorption mechanism”. Journal of the Royal Society, Interface. 12 (109): 20150334. doi:10.1098/rsif.2015.0334. PMC 4535404. PMID 26179989.
- GRCh38: Ensembl release 89: ENSG00000108821 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000001506 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- Sezer S, Şimşek N, Celik HT, Erden G, Ozturk G, Düzgün AP, Çoşkun F, Demircan K (2014). “Association of collagen type I alpha 1 gene polymorphism with inguinal hernia – PubMed”. Hernia: The Journal of Hernias and Abdominal Wall Surgery. 18 (4): 507–12. doi:10.1007/s10029-013-1147-y. PMID 23925543. S2CID 22999363.
- Kawaguchi Y (2018). “Genetic background of degenerative disc disease in the lumbar spine”. Spine Surgery and Related Research. 2 (2): 98–112. doi:10.22603/ssrr.2017-0007. PMC 6698496. PMID 31440655.
- Bode AP, Read MS, Reddick RL (February 1999). “Activation and adherence of lyophilized human platelets on canine vessel strips in the Baumgartner perfusion chamber”. The Journal of Laboratory and Clinical Medicine. 133 (2): 200–211. doi:10.1016/S0022-2143(99)90013-6. PMID 9989772.
- Hollenhorst MA, Tiemeyer KH, Mahoney KE, Aoki K, Ishihara M, Lowery SC, et al. (April 2023). “Comprehensive analysis of platelet glycoprotein Ibα ectodomain glycosylation”. Journal of Thrombosis and Haemostasis. 21 (4): 995–1009. doi:10.1016/j.jtha.2023.01.009. PMC 10065957. PMID 36740532.
- McPherson RA, Pincus MR (2007). Henry’s Clinical Diagnosis and Management by Laboratory Methods (21st ed.). Philadelphia, Pa: Saunders Elsevier. pp. 760–762. ISBN 978-1-4160-0287-1.
- McMillan R (October 2007). “The pathogenesis of chronic immune thrombocytopenic purpura”. Seminars in Hematology. 44 (4 Suppl 5): S3–S11. doi:10.1053/j.seminhematol.2007.11.002. PMID 18096470.
- Li R, Emsley J (April 2013). “The organizing principle of the platelet glycoprotein Ib-IX-V complex”. J. Thromb. Haemost. 11 (4): 605–14. doi:10.1111/jth.12144. PMC 3696474. PMID 23336709.
- McEwan PA, Andrews RK, Emsley J (November 2009). “Glycoprotein Ibalpha inhibitor complex structure reveals a combined steric and allosteric mechanism of von Willebrand factor antagonism”. Blood. 114 (23): 4883–5. doi:10.1182/blood-2009-05-224170. PMID 19726719.
- López JA, Andrews RK, Afshar-Kharghan V, Berndt MC (June 1998). “Bernard-Soulier syndrome”. Blood. 91 (12): 4397–418. doi:10.1182/blood.V91.12.4397. PMID 9616133.
- Du X, Beutler L, Ruan C, Castaldi PA, Berndt MC (May 1987). “Glycoprotein Ib and glycoprotein IX are fully complexed in the intact platelet membrane”. Blood. 69 (5): 1524–7. doi:10.1182/blood.V69.5.1524.1524. PMID 2436691.
- Luo SZ, Mo X, Afshar-Kharghan V, Srinivasan S, López JA, Li R (January 2007). “Glycoprotein Ibalpha forms disulfide bonds with 2 glycoprotein Ibbeta subunits in the resting platelet”. Blood. 109 (2): 603–9. doi:10.1182/blood-2006-05-024091. PMC 1785083. PMID 17008541.
- Mo X, Liu L, López JA, Li R (September 2012). “Transmembrane domains are critical to the interaction between platelet glycoprotein V and glycoprotein Ib-IX complex”. J. Thromb. Haemost. 10 (9): 1875–86. doi:10.1111/j.1538-7836.2012.04841.x. PMC 3499136. PMID 22759073.
- McEwan PA, Yang W, Carr KH, et al. (November 2011). “Quaternary organization of GPIb-IX complex and insights into Bernard–Soulier syndrome revealed by the structures of GPIbβ and a GPIbβ/GPIX chimera”. Blood. 118 (19): 5292–301. doi:10.1182/blood-2011-05-356253. PMC 3217411. PMID 21908432.
- Lanza F (2006). “Bernard-Soulier syndrome (hemorrhagiparous thrombocytic dystrophy)”. Orphanet J Rare Dis. 1: 46. doi:10.1186/1750-1172-1-46. PMC 1660532. PMID 17109744.
- Nurden AT (August 2005). “Qualitative disorders of platelets and megakaryocytes”. J. Thromb. Haemost. 3 (8): 1773–82. doi:10.1111/j.1538-7836.2005.01428.x. PMID 16102044.
- Hollenhorst, Marie A.; Tiemeyer, Katherine H.; Mahoney, Keira E.; Aoki, Kazuhiro; Ishihara, Mayumi; Lowery, Sarah C.; Rangel-Angarita, Valentina; Bertozzi, Carolyn R.; Malaker, Stacy A. (April 2023). “Comprehensive analysis of platelet glycoprotein Ibα ectodomain glycosylation”. Journal of Thrombosis and Haemostasis. 21 (4): 995–1009. doi:10.1016/j.jtha.2023.01.009. PMC 10065957. PMID 36740532.
- Bernard J, Soulier JP (1948). “Sur une nouvelle variete de dystrophie thrombocythaire hemorragipare congenitale”. Sem Hop Paris. 24: 3217–3223.
- Strassel C, David T, Eckly A, et al. (January 2006). “Synthesis of GPIb beta with novel transmembrane and cytoplasmic sequences in a Bernard–Soulier patient resulting in GPIb-defective signaling in CHO cells”. J. Thromb. Haemost. 4 (1): 217–28. doi:10.1111/j.1538-7836.2005.01654.x. PMID 16409472.
- GRCh38: Ensembl release 89: ENSG00000198734 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000026579 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “F5 gene: MedlinePlus Genetics”. medlineplus.gov. Retrieved 2023-03-25.
- Villoutreix BO, Dahlbäck B (June 1998). “Structural investigation of the A domains of human blood coagulation factor V by molecular modeling”. Protein Science. 7 (6): 1317–25. doi:10.1002/pro.5560070607. PMC 2144041. PMID 9655335.
- Thorelli E, Kaufman RJ, Dahlbäck B (June 1998). “The C-terminal region of the factor V B-domain is crucial for the anticoagulant activity of factor V”. The Journal of Biological Chemistry. 273 (26): 16140–45. doi:10.1074/jbc.273.26.16140. PMID 9632668.
- Macedo-Ribeiro S, Bode W, Huber R, Quinn-Allen MA, Kim SW, Ortel TL, Bourenkov GP, Bartunik HD, Stubbs MT, Kane WH, Fuentes-Prior P (November 1999). “Crystal structures of the membrane-binding C2 domain of human coagulation factor V”. Nature. 402 (6760): 434–39. Bibcode:1999Natur.402..434M. doi:10.1038/46594. PMID 10586886. S2CID 4393638.
- Lam W, Moosavi L (18 July 2022). “Physiology, Factor V”. StatPearls. StatPearls Publishing. PMID 31334957. Retrieved 7 February 2022.
- Huang JN, Koerper MA (November 2008). “Factor V deficiency: a concise review”. Haemophilia. 14 (6): 1164–69. doi:10.1111/j.1365-2516.2008.01785.x. PMID 19141156.
- Castoldi E (July 2021). “F5-Atlanta: Factor V-short strikes again”. Journal of Thrombosis and Haemostasis. 19 (7): 1638–1640. doi:10.1111/jth.15351. PMC 8362210. PMID 34176223.
- Stormorken H (February 2003). “The discovery of factor V: a tricky clotting factor”. Journal of Thrombosis and Haemostasis. 1 (2): 206–13. doi:10.1046/j.1538-7836.2003.00043.x. PMID 12871488.
- Owren PA (April 1947). “Parahaemophilia; haemorrhagic diathesis due to absence of a previously unknown clotting factor”. Lancet. 1 (6449): 446–48. doi:10.1016/S0140-6736(47)91941-7. PMID 20293060.
- Jenny RJ, Pittman DD, Toole JJ, Kriz RW, Aldape RA, Hewick RM, Kaufman RJ, Mann KG (July 1987). “Complete cDNA and derived amino acid sequence of human factor V”. Proceedings of the National Academy of Sciences of the United States of America. 84 (14): 4846–50. Bibcode:1987PNAS…84.4846J. doi:10.1073/pnas.84.14.4846. PMC 305202. PMID 3110773
- Bertina RM, Koeleman BP, Koster T, Rosendaal FR, Dirven RJ, de Ronde H, van der Velden PA, Reitsma PH (May 1994). “Mutation in blood coagulation factor V associated with resistance to activated protein C”. Nature. 369 (6475): 64–67. Bibcode:1994Natur.369…64B. doi:10.1038/369064a0. PMID 8164741. S2CID 4314040.
- Heeb MJ, Kojima Y, Rosing J, Tans G, Griffin JH (December 1999). “C-terminal residues 621–635 of protein S are essential for binding to factor Va”. The Journal of Biological Chemistry. 274 (51): 36187–92. doi:10.1074/jbc.274.51.36187. PMID 10593904.
- Heeb MJ, Mesters RM, Tans G, Rosing J, Griffin JH (February 1993). “Binding of protein S to factor Va associated with inhibition of prothrombinase that is independent of activated protein C”. The Journal of Biological Chemistry. 268 (4): 2872–77. doi:10.1016/S0021-9258(18)53854-0. PMID 8428962.
- GRCh38: Ensembl release 89: ENSG00000184500 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000022912 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- Lundwall A, Dackowski W, Cohen E, Shaffer M, Mahr A, Dahlbäck B, Stenflo J, Wydro R (September 1986). “Isolation and sequence of the cDNA for human protein S, a regulator of blood coagulation”. Proc. Natl. Acad. Sci. U.S.A. 83 (18): 6716–20. Bibcode:1986PNAS…83.6716L. doi:10.1073/pnas.83.18.6716. PMC 386580. PMID 2944113.
- Long GL, Marshall A, Gardner JC, Naylor SL (January 1988). “Genes for human vitamin K-dependent plasma proteins C and S are located on chromosomes 2 and 3, respectively”. Somat. Cell Mol. Genet. 14 (1): 93–8. doi:10.1007/BF01535052. PMID 2829367. S2CID 31236887.
- “Protein S deficiency”. UpToDate. Retrieved May 10, 2017.
- Kaushansky K, Lichtman M, Prchal J, Levi M, Press O, Burns L, Caligiuri M (2015). Williams Hematology. McGraw-Hill. p. 1926.
- Stenflo J (1999). “Contributions of Gla and EGF-like domains to the function of vitamin K-dependent coagulation factors”. Critical Reviews in Eukaryotic Gene Expression. 9 (1): 59–88. doi:10.1615/CritRevEukaryotGeneExpr.v9.i1.50. PMID 10200912.
- Rosner W (Dec 1991). “Plasma steroid-binding proteins”. Endocrinology and Metabolism Clinics of North America. 20 (4): 697–720. doi:10.1016/S0889-8529(18)30240-8. PMID 1778174.
- Dahlbäck B, Lundwall A, Stenflo J (Jun 1986). “Primary structure of bovine vitamin K-dependent protein S”. Proceedings of the National Academy of Sciences. 83 (12): 4199–203. Bibcode:1986PNAS…83.4199D. doi:10.1073/pnas.83.12.4199. PMC 323699. PMID 2940598.
- Castoldi E, Hackeng TM (September 2008). “Regulation of coagulation by protein S”. Curr. Opin. Hematol. 15 (5): 529–36. doi:10.1097/MOH.0b013e328309ec97. PMID 18695379. S2CID 11522770.
- Beauchamp NJ, Dykes AC, Parikh N, Campbell Tait R, Daly ME (June 2004). “The prevalence of, and molecular defects underlying, inherited protein S deficiency in the general population”. Br. J. Haematol. 125 (5): 647–54. doi:10.1111/j.1365-2141.2004.04961.x. PMID 15147381. S2CID 705661.
- García de Frutos P, Fuentes-Prior P, Hurtado B, Sala N (September 2007). “Molecular basis of protein S deficiency”. Thromb. Haemost. 98 (3): 543–56. doi:10.1160/th07-03-0199. PMID 17849042. S2CID 17274778.
- Heeb MJ, Kojima Y, Rosing J, Tans G, Griffin J H (Dec 1999). “C-terminal residues 621-635 of protein S are essential for binding to factor Va”. J. Biol. Chem. UNITED STATES. 274 (51): 36187–92. doi:10.1074/jbc.274.51.36187. ISSN 0021-9258. PMID 10593904. S2CID 45995946.
- Heeb MJ, Mesters R M, Tans G, Rosing J, Griffin J H (Feb 1993). “Binding of protein S to factor Va associated with inhibition of prothrombinase that is independent of activated protein C”. J. Biol. Chem. UNITED STATES. 268 (4): 2872–7. doi:10.1016/S0021-9258(18)53854-0. ISSN 0021-9258. PMID 8428962.
- GRCh38: Ensembl release 89: ENSG00000011422 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000046223 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- Kessler, Pascal; Marchot, Pascale; Silva, Marcela; Servent, Denis (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142: 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- Llinas P, Le Du MH, Gårdsvoll H, Danø K, Ploug M, Gilquin B, Stura EA, Ménez A (May 2005). “Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide”. The EMBO Journal. 24 (9): 1655–63. doi:10.1038/sj.emboj.7600635. PMC 1142576. PMID 15861141.
- Huai Q, Mazar AP, Kuo A, Parry GC, Shaw DE, Callahan J, Li Y, Yuan C, Bian C, Chen L, Furie B, Furie BC, Cines DB, Huang M (February 2006). “Structure of human urokinase plasminogen activator in complex with its receptor”. Science. 311 (5761): 656–9. Bibcode:2006Sci…311..656H. doi:10.1126/science.1121143. PMID 16456079. S2CID 39521660.
- ViroGates. “What is suPAR”. suPARnostic® by ViroGates. Retrieved 2021-09-27.
- Thunø M, Macho B, Eugen-Olsen J (2009). “suPAR: the molecular crystal ball”. Disease Markers. 27 (3): 157–72. doi:10.1155/2009/504294. PMC 3835059. PMID 19893210.
- Desmedt S, Desmedt V, Delanghe JR, Speeckaert R, Speeckaert MM (2017). “The Intriguing Role of Soluble Urokinase Receptor in Inflammatory Diseases”. Critical Reviews in Clinical Laboratory Sciences. 54 (2): 117–133. doi:10.1080/10408363.2016.1269310. PMID 28084848. S2CID 32624995.
- Wagner V, Gil J (2020). “T Cells Engineered to Target Senescence”. Nature. 583 (7814): 37–38. Bibcode:2020Natur.583…37W. doi:10.1038/d41586-020-01759-x. hdl:10044/1/80980. PMID 32601490.
- Amor C, Feucht J, Lowe SW (2020). “Senolytic CAR T cells reverse senescence-associated pathologies”. Nature. 583 (7814): 127–132. doi:10.1038/s41586-020-2403-9. PMC 7583560. PMID 32555459.
- Josip Madunić (2018). “The Urokinase Plasminogen Activator System in Human Cancers: An Overview of Its Prognostic and Predictive Role”. Thrombosis and Haemostasis. 118 (12): 2020–2036. doi:10.1055/s-0038-1675399. PMID 30419600.
- Czekay RP, Kuemmel TA, Orlando RA, Farquhar MG (May 2001). “Direct binding of occupied urokinase receptor (uPAR) to LDL receptor-related protein is required for endocytosis of uPAR and regulation of cell surface urokinase activity”. Molecular Biology of the Cell. 12 (5): 1467–79. doi:10.1091/mbc.12.5.1467. PMC 34598. PMID 11359936.
Further reading
- Alvarez-Hernandez D, Naves M, Diaz-Lopez JB, Gomez C, Santamaria I, Cannata-Andia JB (2003). “Influence of polymorphisms in VDR and COLIA1 genes on the risk of osteoporotic fractures in aged men”. Kidney Int Suppl. 63 (85): S14–8. doi:10.1046/j.1523-1755.63.s85.5.x. PMID 12753258.
- Bou-Gharios G, Ponticos M, Rajkumar V, Abraham D (2004). “Extra-cellular matrix in vascular networks”. Cell Proliferation. 37 (3): 207–20. doi:10.1111/j.1365-2184.2004.00306.x. PMC 6496925. PMID 15144498.
- Chamberlain JR, Schwarze U, Wang PR, Hirata RK, Hankenson KD, Pace JM, Underwood RA, Song KM, Sussman M, Byers PH, Russell DW (2004). “Gene targeting in stem cells from individuals with osteogenesis imperfecta”. Science. 303 (5661): 1198–201. Bibcode:2004Sci…303.1198C. doi:10.1126/science.1088757. PMID 14976317. S2CID 13212242.
- Forlino A, Marini JC (2000). “Osteogenesis imperfecta: prospects for molecular therapeutics”. Mol Genet Metab. 71 (1–2): 225–32. doi:10.1006/mgme.2000.3039. PMID 11001814.
- Karsenty G, Park RW (1995). “Regulation of type I collagen genes expression”. Int Rev Immunol. 12 (2–4): 177–85. doi:10.3109/08830189509056711. PMID 7650420.
- Grant SF, Reid DM, Blake G, Herd R, Fogelman I, Ralston SH (1996). “Reduced bone density and osteoporosis associated with a polymorphic Sp1 binding site in the collagen type I alpha 1 gene”. Nat. Genet. 14 (2): 203–05. doi:10.1038/ng1096-203. PMID 8841196. S2CID 20896127.
- Keen RW, Woodford-Richens KL, Grant SF, Ralston SH, Lanchbury JS, Spector TD (1999). “Association of polymorphism at the type I collagen (COL1A1) locus with reduced bone mineral density, increased fracture risk, and increased collagen turnover”. Arthritis Rheum. 42 (2): 285–90. doi:10.1002/1529-0131(199902)42:2<285::AID-ANR10>3.0.CO;2-3. PMID 10025922.
- Kocher MS, Shapiro F (1998). “Osteogenesis imperfecta”. J Am Acad Orthop Surg. 6 (4): 225–36. doi:10.5435/00124635-199807000-00004. PMID 9682085.
- Mann V, Ralston SH (2003). “Meta-analysis of COL1A1 Sp1 polymorphism in relation to bone mineral density and osteoporotic fracture”. Bone. 32 (6): 711–7. doi:10.1016/S8756-3282(03)00087-5. PMID 12810179.
- Nuytinck L, Freund M, Lagae L, Pierard GE, Hermanns-Le T, De Paepe A (2000). “Classical Ehlers-Danlos syndrome caused by a mutation in type I collagen”. Am J Hum Genet. 66 (4): 1398–402. doi:10.1086/302859. PMC 1288203. PMID 10739762.
- Persikov AV, Pillitteri RJ, Amin P, Schwarze U, Byers PH, Brodsky B (2004). “Stability related bias in residues replacing glycines within the collagen triple helix (Gly-Xaa-Yaa) in inherited connective tissue disorders”. Hum Mutat. 24 (4): 330–7. doi:10.1002/humu.20091. PMID 15365990. S2CID 34854398.
- Ralston SH (2002). “Genetic control of susceptibility to osteoporosis”. J Clin Endocrinol Metab. 87 (6): 2460–6. doi:10.1210/jc.87.6.2460. PMID 12050200.
- Byers PH, Wallis GA, Willing MC (1991). “Osteogenesis imperfecta: translation of mutation to phenotype”. J. Med. Genet. 28 (7): 433–42. doi:10.1136/jmg.28.7.433. PMC 1016951. PMID 1895312.
- Kuivaniemi H, Tromp G, Prockop DJ (1991). “Mutations in collagen genes: causes of rare and some common diseases in humans”. FASEB J. 5 (7): 2052–60. doi:10.1096/fasebj.5.7.2010058. PMID 2010058. S2CID 24461341.
- Kuivaniemi H, Tromp G, Prockop DJ (1997). “Mutations in fibrillar collagens (types I, II, III, and XI), fibril-associated collagen (type IX), and network-forming collagen (type X) cause a spectrum of diseases of bone, cartilage, and blood vessels”. Hum. Mutat. 9 (4): 300–15. doi:10.1002/(SICI)1098-1004(1997)9:4<300::AID-HUMU2>3.0.CO;2-9. PMID 9101290. S2CID 6890740.
- Rossert J, Terraz C, Dupont S (2001). “Regulation of type I collagen genes expression”. Nephrol. Dial. Transplant. 15 (Suppl 6): 66–8. doi:10.1093/ndt/15.suppl_6.66. PMID 11143996.
- Nicolaes GA, Dahlbäck B (April 2002). “Factor V and thrombotic disease: description of a janus-faced protein”. Arteriosclerosis, Thrombosis, and Vascular Biology. 22 (4): 530–38. doi:10.1161/01.ATV.0000012665.51263.B7. PMID 11950687.
- Segers K, Dahlbäck B, Nicolaes GA (September 2007). “Coagulation factor V and thrombophilia: background and mechanisms”. Thrombosis and Haemostasis. 98 (3): 530–42. doi:10.1160/th07-02-0150. PMID 17849041. S2CID 29406966.
- Hooper WC, De Staercke C (2006). “The relationship between FV Leiden and pulmonary embolism”. Respiratory Research. 3 (1): 8. doi:10.1186/rr180. PMC 64819. PMID 11806843.
- Schrijver I, Houissa-Kastally R, Jones CD, Garcia KC, Zehnder JL (February 2002). “Novel factor V C2-domain mutation (R2074H) in two families with factor V deficiency and bleeding” (PDF). Thrombosis and Haemostasis. 87 (2): 294–99. doi:10.1055/s-0037-1612988. PMID 11858490. S2CID 4818866. Archived from the original (PDF) on 2019-03-07.
- Mann KG, Kalafatis M (January 2003). “Factor V: a combination of Dr Jekyll and Mr Hyde”. Blood. 101 (1): 20–30. doi:10.1182/blood-2002-01-0290. PMID 12393635. S2CID 8885302.
- Duga S, Asselta R, Tenchini ML (August 2004). “Coagulation factor V”. The International Journal of Biochemistry & Cell Biology. 36 (8): 1393–99. doi:10.1016/j.biocel.2003.08.002. PMID 15147718. S2CID 27193482.
- Andreassi MG, Botto N, Maffei S (2006). “Factor V Leiden, prothrombin G20210A substitution and hormone therapy: indications for molecular screening”. Clinical Chemistry and Laboratory Medicine. 44 (5): 514–21. doi:10.1515/CCLM.2006.103. PMID 16681418. S2CID 34399027.
- Du X (May 2007). “Signaling and regulation of the platelet glycoprotein Ib-IX-V complex”. Current Opinion in Hematology. 14 (3): 262–69. doi:10.1097/MOH.0b013e3280dce51a. PMID 17414217. S2CID 39904506.
- Kunishima S, Kamiya T, Saito H (2003). “Genetic abnormalities of Bernard-Soulier syndrome”. Int. J. Hematol. 76 (4): 319–27. doi:10.1007/BF02982690. PMID 12463594. S2CID 23635810.
- Hickey MJ, Deaven LL, Roth GJ (1991). “Human platelet glycoprotein IX. Characterization of cDNA and localization of the gene to chromosome 3”. FEBS Lett. 274 (1–2): 189–92. doi:10.1016/0014-5793(90)81361-Q. PMID 2253772. S2CID 12468752.
- Du X, Beutler L, Ruan C, et al. (1987). “Glycoprotein Ib and glycoprotein IX are fully complexed in the intact platelet membrane”. Blood. 69 (5): 1524–7. doi:10.1182/blood.V69.5.1524.1524. PMID 2436691.
- Andrews RK, Booth WJ, Gorman JJ, et al. (1990). “Purification of botrocetin from Bothrops jararaca venom. Analysis of the botrocetin-mediated interaction between von Willebrand factor and the human platelet membrane glycoprotein Ib-IX complex”. Biochemistry. 28 (21): 8317–26. doi:10.1021/bi00447a009. PMID 2557900.
- Hickey MJ, Williams SA, Roth GJ (1989). “Human platelet glycoprotein IX: an adhesive prototype of leucine-rich glycoproteins with flank-center-flank structures”. Proc. Natl. Acad. Sci. U.S.A. 86 (17): 6773–7. Bibcode:1989PNAS…86.6773H. doi:10.1073/pnas.86.17.6773. PMC 297928. PMID 2771955
- Roth GJ, Ozols J, Nugent DJ, Williams SA (1988). “Isolation and characterization of human platelet glycoprotein IX”. Biochem. Biophys. Res. Commun. 156 (2): 931–9. doi:10.1016/S0006-291X(88)80933-1. PMID 3056407.
- Meyer SC, Fox JE (1995). “Interaction of platelet glycoprotein V with glycoprotein Ib-IX regulates expression of the glycoproteins and binding of von Willebrand factor to glycoprotein Ib-IX in transfected cells”. J. Biol. Chem. 270 (24): 14693–9. doi:10.1074/jbc.270.24.14693. PMID 7782333.
- Clemetson JM, Kyrle PA, Brenner B, Clemetson KJ (1994). “Variant Bernard-Soulier syndrome associated with a homozygous mutation in the leucine-rich domain of glycoprotein IX”. Blood. 84 (4): 1124–31. doi:10.1182/blood.V84.4.1124.1124. PMID 8049428.
- Hickey MJ, Roth GJ (1993). “Characterization of the gene encoding human platelet glycoprotein IX”. J. Biol. Chem. 268 (5): 3438–43. doi:10.1016/S0021-9258(18)53713-3. PMID 8429020.
- Wright SD, Michaelides K, Johnson DJ, et al. (1993). “Double heterozygosity for mutations in the platelet glycoprotein IX gene in three siblings with Bernard-Soulier syndrome”. Blood. 81 (9): 2339–47. doi:10.1182/blood.V81.9.2339.2339. PMID 8481514.
- Berger G, Massé JM, Cramer EM (1996). “Alpha-granule membrane mirrors the platelet plasma membrane and contains the glycoproteins Ib, IX, and V”. Blood. 87 (4): 1385–95. doi:10.1182/blood.V87.4.1385.bloodjournal8741385. PMID 8608228.
- Hollmann C, Haag F, Schlott M, et al. (1996). “Molecular characterization of mouse T-cell ecto-ADP-ribosyltransferase Rt6: cloning of a second functional gene and identification of the Rt6 gene products”. Mol. Immunol. 33 (9): 807–17. doi:10.1016/0161-5890(96)00008-9. PMID 8811076.
- Noris P, Simsek S, Stibbe J, von dem Borne AE (1997). “A phenylalanine-55 to serine amino-acid substitution in the human glycoprotein IX leucine-rich repeat is associated with Bernard-Soulier syndrome”. Br. J. Haematol. 97 (2): 312–20. doi:10.1046/j.1365-2141.1997.582706.x. PMID 9163595. S2CID 19759549.
- Hayashi T, Suzuki K, Yahagi A, et al. (1997). “Corrected DNA sequence of the platelet glycoprotein IX gene”. Thromb. Haemost. 77 (5): 1034–5. doi:10.1055/s-0038-1656099. PMID 9184424. S2CID 6328233.
- Bradford HN, Dela Cadena RA, Kunapuli SP, et al. (1997). “Human kininogens regulate thrombin binding to platelets through the glycoprotein Ib-IX-V complex”. Blood. 90 (4): 1508–15. doi:10.1182/blood.V90.4.1508. PMID 9269768.
- Suzuki K, Hayashi T, Yahagi A, et al. (1998). “Novel point mutation in the leucine-rich motif of the platelet glycoprotein IX associated with Bernard-Soulier syndrome”. Br. J. Haematol. 99 (4): 794–800. doi:10.1046/j.1365-2141.1997.4753275.x. PMID 9432024. S2CID 20513918.
- Noris P, Arbustini E, Spedini P, et al. (1999). “A new variant of Bernard-Soulier syndrome characterized by dysfunctional glycoprotein (GP) Ib and severely reduced amounts of GPIX and GPV”. Br. J. Haematol. 103 (4): 1004–13. doi:10.1046/j.1365-2141.1998.01100.x. PMID 9886312. S2CID 7435503.
- Longhurst CM, White MM, Wilkinson DA, Jennings LK (1999). “A CD9, alphaIIbbeta3, integrin-associated protein, and GPIb/V/IX complex on the surface of human platelets is influenced by alphaIIbbeta3 conformational states”. Eur. J. Biochem. 263 (1): 104–11. doi:10.1046/j.1432-1327.1999.00467.x. PMID 10429193.
- Kunishima S, Tomiyama Y, Honda S, et al. (2000). “Cys97–>Tyr mutation in the glycoprotein IX gene associated with Bernard-Soulier syndrome”. Br. J. Haematol. 107 (3): 539–45. doi:10.1046/j.1365-2141.1999.01733.x. PMID 10583255. S2CID 28086704.
- Rivera CE, Villagra J, Riordan M, et al. (2001). “Identification of a new mutation in platelet glycoprotein IX (GPIX) in a patient with Bernard-Soulier syndrome”. Br. J. Haematol. 112 (1): 105–8. doi:10.1046/j.1365-2141.2001.02529.x. PMID 11167791. S2CID 887208.
- Dahlbäck B (1991). “Protein S and C4b-binding protein: components involved in the regulation of the protein C anticoagulant system”. Thromb. Haemost. 66 (1): 49–61. doi:10.1055/s-0038-1646373. PMID 1833851. S2CID 24929072.
- Witt I (2002). “Molekularbiologische Grundlagen und Diagnostik der hereditären Defekte von Antithrombin III, Protein C und Protein S” [Molecular biological basis and diagnosis of hereditary defect of antithrombin III, protein c and protein S]. Hamostaseologie (in German). 22 (2): 57–66. doi:10.1055/s-0037-1619540. PMID 12193972. S2CID 58077740.
- Rezende SM, Simmonds RE, Lane DA (2004). “Coagulation, inflammation, and apoptosis: different roles for protein S and the protein S-C4b binding protein complex”. Blood. 103 (4): 1192–201. doi:10.1182/blood-2003-05-1551. PMID 12907438. S2CID 133028.
- Dahlbäck B (2007). “The tale of protein S and C4b-binding protein, a story of affection”. Thromb. Haemost. 98 (1): 90–6. doi:10.1160/th07-04-0269. PMID 17597997. S2CID 18823655.
- García de Frutos P, Fuentes-Prior P, Hurtado B, Sala N (2007). “Molecular basis of protein S deficiency”. Thromb. Haemost. 98 (3): 543–56. doi:10.1160/th07-03-0199. PMID 17849042. S2CID 17274778.
- Maillard C, Berruyer M, Serre CM, et al. (1992). “Protein-S, a vitamin K-dependent protein, is a bone matrix component synthesized and secreted by osteoblasts”. Endocrinology. 130 (3): 1599–604. doi:10.1210/endo.130.3.1531628. PMID 1531628.
- Griffin JH, Gruber A, Fernández JA (1992). “Reevaluation of total, free, and bound protein S and C4b-binding protein levels in plasma anticoagulated with citrate or hirudin”. Blood. 79 (12): 3203–11. doi:10.1182/blood.V79.12.3203.bloodjournal79123203. PMID 1534488.
- Guglielmone HA, Vides MA (1992). “A novel functional assay of protein C in human plasma and its comparison with amidolytic and anticoagulant assays”. Thromb. Haemost. 67 (1): 46–9. doi:10.1055/s-0038-1648377. PMID 1615482. S2CID 27769717.
- Bertina RM, Ploos van Amstel HK, van Wijngaarden A, et al. (1990). “Heerlen polymorphism of protein S, an immunologic polymorphism due to dimorphism of residue 460”. Blood. 76 (3): 538–48. doi:10.1182/blood.V76.3.538.538. PMID 2143091.
- Schmidel DK, Tatro AV, Phelps LG, et al. (1991). “Organization of the human protein S genes”. Biochemistry. 29 (34): 7845–52. doi:10.1021/bi00486a010. PMID 2148110.
- Ploos van Amstel HK, Reitsma PH, van der Logt CP, Bertina RM (1991). “Intron-exon organization of the active human protein S gene PS alpha and its pseudogene PS beta: duplication and silencing during primate evolution”. Biochemistry. 29 (34): 7853–61. doi:10.1021/bi00486a011. PMID 2148111.
- Allaart CF, Aronson DC, Ruys T, et al. (1991). “Hereditary protein S deficiency in young adults with arterial occlusive disease”. Thromb. Haemost. 64 (2): 206–10. PMID 2148653.
- Ohlin AK, Landes G, Bourdon P, et al. (1989). “Beta-hydroxyaspartic acid in the first epidermal growth factor-like domain of protein C. Its role in Ca2+ binding and biological activity”. J. Biol. Chem. 263 (35): 19240–8. doi:10.1016/S0021-9258(18)37415-5. PMID 2461936.
- Schwarz HP, Heeb MJ, Lottenberg R, et al. (1989). “Familial protein S deficiency with a variant protein S molecule in plasma and platelets”. Blood. 74 (1): 213–21. doi:10.1182/blood.V74.1.213.213. PMID 2526663.
- Ploos van Amstel HK, van der Zanden AL, Reitsma PH, Bertina RM (1987). “Human protein S cDNA encodes Phe-16 and Tyr 222 in consensus sequences for the post-translational processing”. FEBS Lett. 222 (1): 186–90. doi:10.1016/0014-5793(87)80217-X. PMID 2820795. S2CID 46365357.
- Dahlbäck B, Lundwall A, Stenflo J (1986). “Primary structure of bovine vitamin K-dependent protein S”. Proc. Natl. Acad. Sci. U.S.A. 83 (12): 4199–203. Bibcode:1986PNAS…83.4199D. doi:10.1073/pnas.83.12.4199. PMC 323699. PMID 2940598.
- Lundwall A, Dackowski W, Cohen E, et al. (1986). “Isolation and sequence of the cDNA for human protein S, a regulator of blood coagulation”. Proc. Natl. Acad. Sci. U.S.A. 83 (18): 6716–20. Bibcode:1986PNAS…83.6716L. doi:10.1073/pnas.83.18.6716. PMC 386580. PMID 2944113.
- Engesser L, Broekmans AW, Briët E, et al. (1987). “Hereditary protein S deficiency: clinical manifestations”. Ann. Intern. Med. 106 (5): 677–82. doi:10.7326/0003-4819-106-5-677. PMID 2952034.
- Watkins PC, Eddy R, Fukushima Y, et al. (1988). “The gene for protein S maps near the centromere of human chromosome 3”. Blood. 71 (1): 238–41. doi:10.1182/blood.V71.1.238.238. PMID 2961379.
- Ploug M (2003). “Structure-function relationships in the interaction between the urokinase-type plasminogen activator and its receptor”. Current Pharmaceutical Design. 9 (19): 1499–528. doi:10.2174/1381612033454630. PMID 12871065.
- Kjøller L (January 2002). “The urokinase plasminogen activator receptor in the regulation of the actin cytoskeleton and cell motility”. Biological Chemistry. 383 (1): 5–19. doi:10.1515/BC.2002.002. PMID 11928822. S2CID 6125978.
- Chavakis T, Kanse SM, May AE, Preissner KT (April 2002). “Haemostatic factors occupy new territory: the role of the urokinase receptor system and kininogen in inflammation”. Biochemical Society Transactions. 30 (2): 168–73. doi:10.1042/BST0300168. PMID 12023845.
- Ploug M, Gårdsvoll H, Jørgensen TJ, Lønborg Hansen L, Danø K (April 2002). “Structural analysis of the interaction between urokinase-type plasminogen activator and its receptor: a potential target for anti-invasive cancer therapy”. Biochemical Society Transactions. 30 (2): 177–83. doi:10.1042/BST0300177. PMID 12023847.
- Alfano M, Sidenius N, Blasi F, Poli G (November 2003). “The role of urokinase-type plasminogen activator (uPA)/uPA receptor in HIV-1 infection”. Journal of Leukocyte Biology. 74 (5): 750–6. doi:10.1189/jlb.0403176. PMID 12960238. S2CID 8526093.
- Alfano D, Franco P, Vocca I, Gambi N, Pisa V, Mancini A, Caputi M, Carriero MV, Iaccarino I, Stoppelli MP (February 2005). “The urokinase plasminogen activator and its receptor: role in cell growth and apoptosis”. Thrombosis and Haemostasis. 93 (2): 205–11. doi:10.1160/TH04-09-0592. PMID 15711734. S2CID 35517406.
External links
- GeneReviews/NCBI/NIH/UW entry on von Willebrand Factor Deficiency. Includes: Type 1 von Willebrand Disease, Type 2A von Willebrand Disease, Type 2B von Willebrand Disease, Type 2M von Willebrand Disease, Type 2N von Willebrand Disease, Type 3 von Willebrand Disease
- Overview of all the structural information available in the PDB for UniProt: P04275 (von Willebrand factor) at the PDBe-KB.

Leave a Reply