Three-finger proteins and three-finger toxins
Three-finger proteins or three-finger protein domains (3FP or TFPD) are a protein superfamily consisting of small, roughly 60-80 amino acid residue protein domains with a common tertiary structure: three beta strand loops extended from a hydrophobic core stabilized by disulfide bonds. The family is named for the outstretched “fingers” of the three loops. Members of the family have no enzymatic activity, but are capable of forming protein-protein interactions with high specificity and affinity. The founding members of the family, also the best characterized by structure, are the three-finger toxins found in snake venom, which have a variety of pharmacological effects, most typically by disruption of cholinergic signaling. The family is also represented in non-toxic proteins, which have a wide taxonomic distribution; 3FP domains occur in the extracellular domains of some cell-surface receptors as well as in GPI-anchored and secreted globular proteins, usually involved in signaling.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Hegde RP, Rajagopalan N, Doley R, Kini M (2010). “Snake venom three-finger toxins”. In Mackessy SP (ed.). Handbook of venoms and toxins of reptiles. Boca Raton: CRC Press. pp. 287–302. ISBN 9781420008661.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- Utkin Y, Sunagar K, Jackson TN, Reeks T, Fry BG (2015). “Chapter 8: Three-finger toxins”. In Fry B (ed.). Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. Oxford University Press. pp. 218–227. ISBN 9780199309405.
Three-finger toxins
The founding members of the 3FP family are the three-finger toxins (3FTx) often found in snake venom. 3FTx proteins are widely distributed in venomous snake families, but are particularly enriched in the family Elapidae, in which the relative proportion of 3FTx to other venom toxins can reach 95%. Many 3FTx proteins are neurotoxins, though the mechanism of toxicity varies significantly even among proteins of relatively high sequence identity; common protein targets include those involved in cholinergic signaling, such as the nicotinic acetylcholine receptors, muscarinic acetylcholine receptors, and acetylcholinesterase. Another large subfamily of 3FTx proteins is the cardiotoxins (also known as cytotoxins or cytolysins); this group is directly cytotoxic most likely due to interactions with phospholipids and possibly other components of the cell membrane.
- Hegde RP, Rajagopalan N, Doley R, Kini M (2010). “Snake venom three-finger toxins”. In Mackessy SP (ed.). Handbook of venoms and toxins of reptiles. Boca Raton: CRC Press. pp. 287–302. ISBN 9781420008661.
- Utkin Y, Sunagar K, Jackson TN, Reeks T, Fry BG (2015). “Chapter 8: Three-finger toxins”. In Fry B (ed.). Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. Oxford University Press. pp. 218–227. ISBN 9780199309405.
- Sanz L, Pla D, Pérez A, Rodríguez Y, Zavaleta A, Salas M, Lomonte B, Calvete JJ (June 2016). “Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA₂ Dichotomy across Micrurus Venoms”. Toxins. 8 (6): 178. doi:10.3390/toxins8060178. PMC 4926144. PMID 27338473.
Main article: Three-finger toxin
Three-finger toxins (abbreviated 3FTx) are a protein superfamily of small toxin proteins found in the venom of snakes. Three-finger toxins are in turn members of a larger superfamily of three-finger protein domains which includes non-toxic proteins that share a similar protein fold. The group is named for its common structure consisting of three beta strand loops connected to a central core containing four conserved disulfide bonds. The 3FP protein domain has no enzymatic activity and is typically between 60-74 amino acid residues long. Despite their conserved structure, three-finger toxin proteins have a wide range of pharmacological effects. Most members of the family are neurotoxins that act on cholinergic intercellular signaling; the alpha-neurotoxin family interacts with muscle nicotinic acetylcholine receptors (nAChRs), the kappa-bungarotoxin family with neuronal nAChRs, and muscarinic toxins with muscarinic acetylcholine receptors (mAChRs).
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Hegde RP, Rajagopalan N, Doley R, Kini M (2010). “Snake venom three-finger toxins”. In Mackessy SP (ed.). Handbook of venoms and toxins of reptiles. Boca Raton: CRC Press. pp. 287–302. ISBN 9781420008661.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- Utkin Y, Sunagar K, Jackson T, Reeks T, Fry BG (2015). “Chapter 8: Three-finger toxins”. In Fry B (ed.). Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. Oxford University Press. pp. 218–227. ISBN 9780199309405.
Structure
The three-finger toxin superfamily is defined by a common tertiary structure consisting of three beta strand-containing loops (designated loops I, II, and III) projecting from a small hydrophobic core containing four conserved disulfide bonds. This structure is thought to resemble a hand with three fingers, giving rise to the name. The proteins are typically 60-74 amino acid residues long, though some have additional N- or C-terminal extensions. An additional disulfide bond may be present in either loop I or loop II. The superfamily can be broadly divided into three classes:
- short-chain toxins have under 66 residues and four core disulfide bonds.
- long-chain toxins have at least 66 residues, a disulfide bond in loop II, and possibly a C-terminal extension.
- non-conventional toxins have a disulfide bond in loop I and possibly terminal extensions.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Hegde RP, Rajagopalan N, Doley R, Kini M (2010). “Snake venom three-finger toxins”. In Mackessy SP (ed.). Handbook of venoms and toxins of reptiles. Boca Raton: CRC Press. pp. 287–302. ISBN 9781420008661.
- “Snake three-finger toxin family”. VenomZone. Retrieved 21 April 2017.
Oligomerization
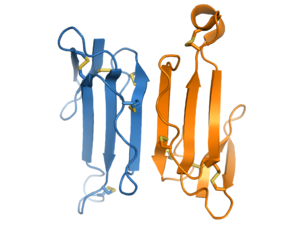
Most 3FTx proteins are monomers. However, some 3FTx subgroups form functional non-covalent homodimers. The kappa-bungarotoxin group is the best characterized dimeric 3FTx, and interacts through an antiparallel dimer interface composed of the outer strand of loop III. Haditoxin is another example of a dimeric 3FTx; it is a member of the short-chain group and has a similar dimer interface but distinct pharmacology compared to the long-chain-like kappa-bungarotoxins.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Dewan JC, Grant GA, Sacchettini JC (November 1994). “Crystal structure of kappa-bungarotoxin at 2.3-A resolution”. Biochemistry. 33 (44): 13147–54. doi:10.1021/bi00248a026. PMID 7947721.
A few examples of covalently linked dimers have also been described. These proteins, from the non-conventional group, are linked through intermolecular disulfide bonds. Some, such as irditoxin, are heterodimers linked by cysteines in loops I and II. Others, such as α-cobratoxin, can form both homodimers and heterodimers which have distinct pharmacological activities in vitro, though their functional significance is unclear due to their very low concentration in venom.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Osipov AV, Kasheverov IE, Makarova YV, Starkov VG, Vorontsova OV, Ziganshin RK, Andreeva TV, Serebryakova MV, Benoit A, Hogg RC, Bertrand D, Tsetlin VI, Utkin YN (May 2008). “Naturally occurring disulfide-bound dimers of three-fingered toxins: a paradigm for biological activity diversification”. The Journal of Biological Chemistry. 283 (21): 14571–80. doi:10.1074/jbc.M802085200. PMID 18381281.
- Pawlak J, Mackessy SP, Sixberry NM, Stura EA, Le Du MH, Ménez R, Foo CS, Ménez A, Nirthanan S, Kini RM (February 2009). “Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity”. FASEB Journal. 23 (2): 534–45. doi:10.1096/fj.08-113555. PMID 18952712. S2CID 4816592.
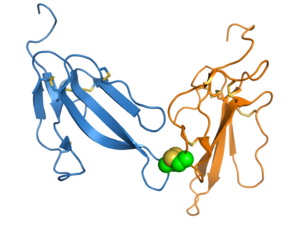
Function
Despite their conserved shared structure, 3FTx proteins have a wide range of pharmacological effects mediating their toxicity. Many members of the family are neurotoxins that bind to receptor proteins in the cell membrane, particularly nicotinic acetylcholine receptors. Others, including the second-largest 3FTx subgroup, are cardiotoxins.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
Cellular targets
Nicotinic acetylcholine receptors
Many of the most well-characterized 3FTx proteins exert their toxic effects through binding to nicotinic acetylcholine receptors (nAChRs), a family of ligand-gated ion channels. 3FTx binding interferes with cholinergic intercellular signaling particularly at neuromuscular junctions and causes paralysis. The alpha-neurotoxin family is a group of 3FTx proteins that bind muscle nAChRs, preventing the binding of the neurotransmitter acetylcholine. Alpha-bungarotoxin, the alpha-neurotoxin from the many-banded krait (Bungarus multicinctus), has a long history of use in molecular biology research; it was through the study of this toxin that nAChRs were isolated and characterized, which facilitated the study of the subunit composition of tissue-specific nAChRs and the detailed pharmacological understanding of the neuromuscular junction. In general, short-chain 3FTx members of this group bind muscle nAChRs only, and long-chain members bind both muscle and neuronal receptors. This 3FTx group is sometimes referred to as the “curaremimetic” toxins due to the similarity of their effects with the plant alkaloid curare.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- de la Rosa, Guillermo; Olvera, Felipe; Archundia, Irving G.; Lomonte, Bruno; Alagón, Alejandro; Corzo, Gerardo (December 2019). “Horse immunization with short-chain consensus α-neurotoxin generates antibodies against broad spectrum of elapid venomous species”. Nature Communications. 10 (1): 3642. Bibcode:2019NatCo..10.3642D. doi:10.1038/s41467-019-11639-2. ISSN 2041-1723. PMC 6692343. PMID 31409779.
- Nirthanan S, Gwee MC (January 2004). “Three-finger α-neurotoxins and the nicotinic acetylcholine receptor, forty years on”. Journal of Pharmacological Sciences. 94 (1): 1–17. doi:10.1254/jphs.94.1. PMID 14745112.
Other groups of 3FTx proteins also bind to different nAChR subtypes; for example, kappa-neurotoxins, which are long-chain dimers, bind neuronal nAChRs, and haditoxin, which is a short-chain dimer, binds both muscle and neuronal subtypes. Non-conventional 3FTx proteins also often bind nAChRs; these were thought to be weaker toxins when first discovered, but the class has been found to possess a range of binding affinities. Recently, a new class of nAChR antagonist 3FTx proteins called omega-neurotoxins has been described.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Hassan-Puttaswamy V, Adams DJ, Kini RM (December 2015). “A Distinct Functional Site in Ω-Neurotoxins: Novel Antagonists of Nicotinic Acetylcholine Receptors from Snake Venom”. ACS Chemical Biology. 10 (12): 2805–15. doi:10.1021/acschembio.5b00492. PMID 26448325.
Muscarinic acetylcholine receptors
A smaller class of 3FTx proteins binds instead to muscarinic acetylcholine receptors, a family of G-protein-coupled receptors. Muscarinic toxins can be either receptor agonists or receptor antagonists, and in some cases the same 3FTx protein is an agonist at one receptor subtype and an antagonist at another. Muscarinic toxins are generally of the short-chain type.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
Acetylcholinesterase
A class of 3FTx proteins called fasciculins bind the enzyme acetylcholinesterase and inhibit its activity by blocking access of acetylcholine to the enzyme’s active site, thereby preventing acetylcholine breakdown. This class derives its name from its clinical effect, causing muscle fasciculations.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Karlsson E, Mbugua PM, Rodriguez-Ithurralde D (1984-01-01). “Fasciculins, anticholinesterase toxins from the venom of the green mamba Dendroaspis angusticeps”. Journal de Physiologie. 79 (4): 232–40. PMID 6530667.
Cardiac targets
The second-largest class of 3FTx proteins causes toxicity in cardiac myocytes and can cause increased heart rate and eventually cardiac arrest. These cardiotoxins also often have generalized cytotoxic effects and are sometimes known as cytolysins. The protein targets in myocytes are not generally known for this class, though some members may cause physical damage to the cell by establishing pores in the cell membrane.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
Another class, called the beta-cardiotoxins, causes decreased heart rate and are thought to function as beta blockers, antagonists for the beta-1 and beta-2 adrenergic receptors.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Rajagopalan N, Pung YF, Zhu YZ, Wong PT, Kumar PP, Kini RM (November 2007). “Beta-cardiotoxin: a new three-finger toxin from Ophiophagus hannah (king cobra) venom with beta-blocker activity”. FASEB Journal. 21 (13): 3685–95. doi:10.1096/fj.07-8658com. PMID 17616557. S2CID 21235585.
Less common targets
There are known 3FTx proteins that target a variety of additional protein targets to exert their toxic effects. For example, L-type calcium channels are targeted by calciseptine and platelet aggregation is inhibited via interactions with adhesion proteins by dendroaspin and related proteins. In some cases no toxicity is observed as a result of the 3FTx-target interaction; for example, the mambalgin family of 3FTx proteins interacts with acid-sensing ion channels to produce analgesia without apparent toxic effect in laboratory tests.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Diochot S, Baron A, Salinas M, Douguet D, Scarzello S, Dabert-Gay AS, Debayle D, Friend V, Alloui A, Lazdunski M, Lingueglia E (October 2012). “Black mamba venom peptides target acid-sensing ion channels to abolish pain” (PDF). Nature. 490 (7421): 552–5. Bibcode:2012Natur.490..552D. doi:10.1038/nature11494. PMID 23034652. S2CID 4337253.
Orphan 3FTx proteins
Bioinformatics-based surveys of known protein sequences have often identified a number of sequences likely to form a 3FTx protein structure but whose function has not been experimentally characterized. Thus, it is not known whether these “orphan” proteins are in fact toxins or what their cellular targets might be. Genomics studies of gene expression in snakes have shown that members of protein families traditionally considered toxins are widely expressed in snake body tissues and that this expression pattern occurs outside the highly venomous superfamily Caenophidia.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Fry BG, Wüster W, Kini RM, Brusic V, Khan A, Venkataraman D, Rooney AP (July 2003). “Molecular evolution and phylogeny of elapid snake venom three-finger toxins”. Journal of Molecular Evolution. 57 (1): 110–29. Bibcode:2003JMolE..57..110F. CiteSeerX 10.1.1.539.324. doi:10.1007/s00239-003-2461-2. PMID 12962311. S2CID 12358977.
- Reyes-Velasco J, Card DC, Andrew AL, Shaney KJ, Adams RH, Schield DR, Casewell NR, Mackessy SP, Castoe TA (January 2015). “Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom”. Molecular Biology and Evolution. 32 (1): 173–83. doi:10.1093/molbev/msu294. PMID 25338510.
Structure-function activity relationships
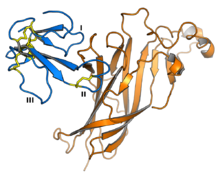
Because 3FTx proteins of similar structure bind a diverse range of cellular protein targets, the relationships between 3FTx protein sequence and their biological activity have been studied extensively, especially among the alpha-neurotoxins. Known functional sites conferring binding affinity and specificity are concentrated in the loops of 3FTx proteins. For example, the crystal structure of alpha-bungarotoxin in complex with the extracellular domain of the alpha-9 nAChR subunit indicates a protein-protein interaction mediated through loops I and II, with no contacts formed by loop III. Interaction surfaces have been mapped for a number of toxins and vary in which loops participate in binding; erabutoxin A uses all three loops to bind nAChRs, while the dendroaspin interaction with adhesion proteins is mediated by three residues in loop III. In some 3FTx proteins with a C-terminal extension, these residues also participate in forming key binding interactions.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Zouridakis M, Giastas P, Zarkadas E, Chroni-Tzartou D, Bregestovski P, Tzartos SJ (November 2014). “Crystal structures of free and antagonist-bound states of human α9 nicotinic receptor extracellular domain”. Nature Structural & Molecular Biology. 21 (11): 976–80. doi:10.1038/nsmb.2900. PMID 25282151. S2CID 30096256.
- Trémeau O, Lemaire C, Drevet P, Pinkasfeld S, Ducancel F, Boulain JC, Ménez A (April 1995). “Genetic engineering of snake toxins. The functional site of Erabutoxin a, as delineated by site-directed mutagenesis, includes variant residues”. The Journal of Biological Chemistry. 270 (16): 9362–9. doi:10.1074/jbc.270.16.9362. PMID 7721859.
- Lu X, Davies J, Lu D, Xia M, Wattam B, Shang D, Sun Y, Scully M, Kakkar V (2006-01-01). “The effect of the single substitution of arginine within the RGD tripeptide motif of a modified neurotoxin dendroaspin on its activity of platelet aggregation and cell adhesion”. Cell Communication & Adhesion. 13 (3): 171–83. doi:10.1080/15419060600726183. PMID 16798616.
The cardiotoxin/cytolysin 3FTx subgroup has a somewhat different set of functionally significant residues due to its distinct mechanism of action, likely involving interactions with phospholipids in the cell membrane, as well as possible functionally significant interactions with other cell-surface molecules such as glycosaminoglycans. A hydrophobic patch of residues contiguous in tertiary structure but distributed over all three loops has been identified as functionally significant in combination with a set of conserved lysine residues conferring local positive charge.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Konshina AG, Boldyrev IA, Utkin YN, Omel’kov AV, Efremov RG (April 2011). “Snake cytotoxins bind to membranes via interactions with phosphatidylserine head groups of lipids”. PLOS ONE. 6 (4): e19064. Bibcode:2011PLoSO…619064K. doi:10.1371/journal.pone.0019064. PMC 3084733. PMID 21559494.
- Lee SC, Lin CC, Wang CH, Wu PL, Huang HW, Chang CI, Wu WG (July 2014). “Endocytotic routes of cobra cardiotoxins depend on spatial distribution of positively charged and hydrophobic domains to target distinct types of sulfated glycoconjugates on cell surface”. The Journal of Biological Chemistry. 289 (29): 20170–81. doi:10.1074/jbc.M114.557157. PMC 4106332. PMID 24898246.
Because of their structural similarity and functional diversity, 3FTx proteins have been used as model systems for the study of protein engineering. Their high binding specificity against targets of pharmacological interest, lack of enzymatic activity, and low immunogenicity have also prompted interest in their potential as drug leads.
- Fruchart-Gaillard C, Mourier G, Blanchet G, Vera L, Gilles N, Ménez R, Marcon E, Stura EA, Servent D (2012-06-14). “Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors”. PLOS ONE. 7 (6): e39166. Bibcode:2012PLoSO…739166F. doi:10.1371/journal.pone.0039166. PMC 3375269. PMID 22720062.
- Georgieva D, Arni RK, Betzel C (December 2008). “Proteome analysis of snake venom toxins: pharmacological insights”. Expert Review of Proteomics. 5 (6): 787–97. doi:10.1586/14789450.5.6.787. PMID 19086859. S2CID 29126350.
- Saviola AJ, Peichoto ME, Mackessy SP (2014-12-01). “Rear-fanged snake venoms: an untapped source of novel compounds and potential drug leads”. Toxin Reviews. 33 (4): 185–201. doi:10.3109/15569543.2014.942040. hdl:11336/17030. ISSN 1556-9543. S2CID 30100277
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
Evolution
Although three-finger proteins in general are widely distributed among metazoans, three-finger toxins appear only in snakes. They are usually considered to be restricted to the Caenophidia lineage (the taxon containing all venomous snakes), though at least one putative 3FTx homolog has been identified in the genome of the Burmese python, a member of a sister taxon. Early work in analyzing protein homology by sequence alignment in the 1970s suggested 3FTx proteins may have evolved from an ancestral ribonuclease; however, more recent molecular phylogeny studies indicate that 3FTx proteins evolved from non-toxic three-finger proteins.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- Strydom, D. J. (December 1973). “Snake Venom Toxins: The Evolution of Some of the Toxins Found in Snake Venoms”. Systematic Zoology. 22 (4): 596–608. doi:10.2307/2412964. JSTOR 2412964.
- Fry BG (March 2005). “From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins”. Genome Research. 15 (3): 403–20. doi:10.1101/gr.3228405. PMC 551567. PMID 15741511.
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG (April 2013). “Complex cocktails: the evolutionary novelty of venoms”. Trends in Ecology & Evolution. 28 (4): 219–29. doi:10.1016/j.tree.2012.10.020. PMID 23219381.
- Fry BG, Wüster W, Kini RM, Brusic V, Khan A, Venkataraman D, Rooney AP (July 2003). “Molecular evolution and phylogeny of elapid snake venom three-finger toxins”. Journal of Molecular Evolution. 57 (1): 110–29. Bibcode:2003JMolE..57..110F. CiteSeerX 10.1.1.539.324. doi:10.1007/s00239-003-2461-2. PMID 12962311. S2CID 12358977.
- Reyes-Velasco J, Card DC, Andrew AL, Shaney KJ, Adams RH, Schield DR, Casewell NR, Mackessy SP, Castoe TA (January 2015). “Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom”. Molecular Biology and Evolution. 32 (1): 173–83. doi:10.1093/molbev/msu294. PMID 25338510.
Among venomous snakes, the distribution of 3FTx proteins varies; they are particularly enriched in venom from the family Elapidae. In the king cobra (Ophiophagus hannah) and Eastern green mamba (Dendroaspis angusticeps), 3FTx proteins make up about 70% of the protein toxins in venom; in the desert coral snake (Micrurus tschudii) the proportion is reported as high as 95%.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- de la Rosa, Guillermo; Corrales-García, Ligia L.; Rodriguez-Ruiz, Ximena; López-Vera, Estuardo; Corzo, Gerardo (July 2018). “Short-chain consensus alpha-neurotoxin: a synthetic 60-mer peptide with generic traits and enhanced immunogenic properties”. Amino Acids. 50 (7): 885–895. doi:10.1007/s00726-018-2556-0. ISSN 0939-4451. PMID 29626299. S2CID 4638613.
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJ, Kerkkamp HM, Vos RA, Guerreiro I, Calvete JJ, Wüster W, Woods AE, Logan JM, Harrison RA, Castoe TA, de Koning AP, Pollock DD, Yandell M, Calderon D, Renjifo C, Currier RB, Salgado D, Pla D, Sanz L, Hyder AS, Ribeiro JM, Arntzen JW, van den Thillart GE, Boetzer M, Pirovano W, Dirks RP, Spaink HP, Duboule D, McGlinn E, Kini RM, Richardson MK (December 2013). “The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system”. Proceedings of the National Academy of Sciences of the United States of America. 110 (51): 20651–6. Bibcode:2013PNAS..11020651V. doi:10.1073/pnas.1314702110. PMC 3870661. PMID 24297900.
- Lauridsen LP, Laustsen AH, Lomonte B, Gutiérrez JM (March 2016). “Toxicovenomics and antivenom profiling of the Eastern green mamba snake (Dendroaspis angusticeps)” (PDF). Journal of Proteomics. 136: 248–61. doi:10.1016/j.jprot.2016.02.003. PMID 26877184.
- Sanz L, Pla D, Pérez A, Rodríguez Y, Zavaleta A, Salas M, Lomonte B, Calvete JJ (June 2016). “Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA₂ Dichotomy across Micrurus Venoms”. Toxins. 8 (6): 178. doi:10.3390/toxins8060178. PMC 4926144. PMID 27338473.
Genes encoding three-finger toxins are thought to have evolved through gene duplication. Traditionally, this has been conceptualized as repeated events of duplication followed by neofunctionalization and recruitment to gene expression patterns restricted to venom glands. However, it has been argued that this process should be extremely rare and that subfunctionalization better explains the observed distribution. More recently, non-toxic 3FP proteins have been found to be widely expressed in many different tissues in snakes, prompting the alternative hypothesis that proteins of restricted expression in saliva were selectively recruited for toxic functionality. There is evidence that most types of 3FTx proteins have been subject to positive selection (that is, diversifying selection) in their recent evolutionary history, possibly due to an evolutionary arms race with prey species. Notable exceptions are the dimeric kappa-bungarotoxin family, likely as a result of evolutionary constraints on the dimer interface, and the cardiotoxin/cytotoxin family, in which a larger fraction of the protein’s residues are believed to have functional roles.
- Fry BG (March 2005). “From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins”. Genome Research. 15 (3): 403–20. doi:10.1101/gr.3228405. PMC 551567. PMID 15741511.
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG (April 2013). “Complex cocktails: the evolutionary novelty of venoms”. Trends in Ecology & Evolution. 28 (4): 219–29. doi:10.1016/j.tree.2012.10.020. PMID 23219381.
- Fry BG, Casewell NR, Wüster W, Vidal N, Young B, Jackson TN (September 2012). “The structural and functional diversification of the Toxicofera reptile venom system”. Toxicon. Advancing in Basic and Translational Venomics. 60 (4): 434–48. doi:10.1016/j.toxicon.2012.02.013. PMID 22446061.
- Hargreaves AD, Swain MT, Hegarty MJ, Logan DW, Mulley JF (August 2014). “Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins”. Genome Biology and Evolution. 6 (8): 2088–95. doi:10.1093/gbe/evu166. PMC 4231632. PMID 25079342.
- Sunagar K, Jackson TN, Undheim EA, Ali SA, Antunes A, Fry BG (November 2013). “Three-fingered RAVERs: Rapid Accumulation of Variations in Exposed Residues of snake venom toxins”. Toxins. 5 (11): 2172–208. doi:10.3390/toxins5112172. PMC 3847720. PMID 24253238.
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJ, Kerkkamp HM, Vos RA, Guerreiro I, Calvete JJ, Wüster W, Woods AE, Logan JM, Harrison RA, Castoe TA, de Koning AP, Pollock DD, Yandell M, Calderon D, Renjifo C, Currier RB, Salgado D, Pla D, Sanz L, Hyder AS, Ribeiro JM, Arntzen JW, van den Thillart GE, Boetzer M, Pirovano W, Dirks RP, Spaink HP, Duboule D, McGlinn E, Kini RM, Richardson MK (December 2013). “The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system”. Proceedings of the National Academy of Sciences of the United States of America. 110 (51): 20651–6. Bibcode:2013PNAS..11020651V. doi:10.1073/pnas.1314702110. PMC 3870661. PMID 24297900.
- Reyes-Velasco J, Card DC, Andrew AL, Shaney KJ, Adams RH, Schield DR, Casewell NR, Mackessy SP, Castoe TA (January 2015). “Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom”. Molecular Biology and Evolution. 32 (1): 173–83. doi:10.1093/molbev/msu294. PMID 25338510.
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJ, Kerkkamp HM, Vos RA, Guerreiro I, Calvete JJ, Wüster W, Woods AE, Logan JM, Harrison RA, Castoe TA, de Koning AP, Pollock DD, Yandell M, Calderon D, Renjifo C, Currier RB, Salgado D, Pla D, Sanz L, Hyder AS, Ribeiro JM, Arntzen JW, van den Thillart GE, Boetzer M, Pirovano W, Dirks RP, Spaink HP, Duboule D, McGlinn E, Kini RM, Richardson MK (December 2013). “The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system”. Proceedings of the National Academy of Sciences of the United States of America. 110 (51): 20651–6. Bibcode:2013PNAS..11020651V. doi:10.1073/pnas.1314702110. PMC 3870661. PMID 24297900.
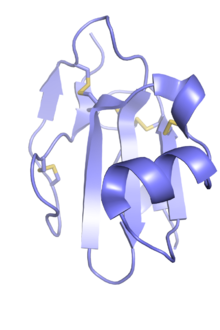
Ly6/uPAR family
TR, Smith RA, Mackenzie L, Morgan BP,
Lea SM (August 2007). “High-resolution
structures of bacterially expressed soluble human CD59″. Acta Crystallographica. Section F, Structural Biology and Crystallization Communications. 63 (Pt 8): 648–52. doi:10.1107/S1744309107033477.
PMC 2335151. PMID 17671359.
The Ly6/uPAR family broadly describes a gene family containing three-finger protein domains that are not toxic and not venom components; these are often known as LU domains and can be found in the extracellular domains of cell-surface receptors and in either GPI-anchored or secreted globular proteins. The family is named for two representative groups of members, the small globular protein lymphocyte antigen 6 (LY6) family and the urokinase plasminogen activator receptor (uPAR). Other receptors with LU domains include members of the transforming growth factor beta receptor (TGF-beta) superfamily, such as the activin type 2 receptor; and bone morphogenetic protein receptor, type IA. Other LU domain proteins are small globular proteins such as CD59 antigen, LYNX1, SLURP1, and SLURP2.
- Loughner CL, Bruford EA, McAndrews MS, Delp EE, Swamynathan S, Swamynathan SK (April 2016). “Organization, evolution and functions of the human and mouse Ly6/uPAR family genes”. Human Genomics. 10: 10. doi:10.1186/s40246-016-0074-2. PMC 4839075. PMID 27098205.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549. ^
- Ploug M, Ellis V (August 1994). “Structure-function relationships in the receptor for urokinase-type plasminogen activator. Comparison to other members of the Ly-6 family and snake venom alpha-neurotoxins”. FEBS Letters. 349 (2): 163–8. doi:10.1016/0014-5793(94)00674-1. PMID 8050560. S2CID 86302713.
- Greenwald J, Fischer WH, Vale WW, Choe S (January 1999). “Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase”. Nature Structural Biology. 6 (1): 18–22. doi:10.1038/4887. PMID 9886286. S2CID 26301441.
- Kirsch T, Sebald W, Dreyer MK (June 2000). “Crystal structure of the BMP-2-BRIA ectodomain complex”. Nature Structural Biology. 7 (6): 492–6. doi:10.1038/75903. PMID 10881198. S2CID 19403233.
- Galat A (November 2008). “The three-fingered protein domain of the human genome”. Cellular and Molecular Life Sciences. 65 (21): 3481–93. doi:10.1007/s00018-008-8473-8. PMID 18821057. S2CID 19931506
Many LU domain containing proteins are involved in cholinergic signaling and bind acetylcholine receptors, notably linking their function to a common mechanism of 3FTx toxicity. Members of the Ly6/uPAR family are believed to be the evolutionary ancestors of 3FTx toxins.
- Tsetlin VI (February 2015). “Three-finger snake neurotoxins and Ly6 proteins targeting nicotinic acetylcholine receptors: pharmacological tools and endogenous modulators”. Trends in Pharmacological Sciences. 36 (2): 109–23. doi:10.1016/j.tips.2014.11.003. PMID 25528970.
- Fry BG (March 2005). “From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins”. Genome Research. 15 (3): 403–20. doi:10.1101/gr.3228405. PMC 551567. PMID 15741511.
- Loughner CL, Bruford EA, McAndrews MS, Delp EE, Swamynathan S, Swamynathan SK (April 2016). “Organization, evolution and functions of the human and mouse Ly6/uPAR family genes”. Human Genomics. 10: 10. doi:10.1186/s40246-016-0074-2. PMC 4839075. PMID 27098205.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
Other LU proteins, such as the CD59 antigen, have well-studied functions in regulation of the immune system.
- Tsetlin VI (February 2015). “Three-finger snake neurotoxins and Ly6 proteins targeting nicotinic acetylcholine receptors: pharmacological tools and endogenous modulators”. Trends in Pharmacological Sciences. 36 (2): 109–23. doi:10.1016/j.tips.2014.11.003. PMID 25528970.
Gene structure
Snake three-finger toxins and the Ly6/uPAR family members share a common gene structure, typically consisting of two introns and three exons. The sequence of the first exon is generally well conserved compared to the other two. The third exon contains the major differentiating features between the two groups, as this is where the C-terminal GPI-anchor peptide common among the Ly6/uPAR globular proteins is encoded.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- Tsetlin VI (February 2015). “Three-finger snake neurotoxins and Ly6 proteins targeting nicotinic acetylcholine receptors: pharmacological tools and endogenous modulators”. Trends in Pharmacological Sciences. 36 (2): 109–23. doi:10.1016/j.tips.2014.11.003. PMID 25528970.
Evolution and taxonomic distribution
Proteins of the general three-finger fold are widely distributed among metazoans. A 2008 bioinformatics study identified about 45 examples of such proteins, containing up to three three-finger domains, represented in the human genome. A more recent profile of the Ly6/uPAR gene family identified 35 human and at least 61 mouse family members in the organisms’ respective genomes.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- Galat A (November 2008). “The three-fingered protein domain of the human genome”. Cellular and Molecular Life Sciences. 65 (21): 3481–93. doi:10.1007/s00018-008-8473-8. PMID 18821057. S2CID 19931506.
- Loughner CL, Bruford EA, McAndrews MS, Delp EE, Swamynathan S, Swamynathan SK (April 2016). “Organization, evolution and functions of the human and mouse Ly6/uPAR family genes”. Human Genomics. 10: 10. doi:10.1186/s40246-016-0074-2. PMC 4839075. PMID 27098205
The three-finger protein family is thought to have expanded through gene duplication in the snake lineage. 3FTx toxins are considered restricted to the Caenophidia, the taxon containing all venomous snakes; however at least one homolog has been identified in the Burmese python, a closely related subgroup. Traditionally, 3FTx genes have been thought to have evolved by repeated events of duplication followed by neofunctionalization and recruitment to gene expression patterns restricted to venom glands. However, it has been argued that this process should be extremely rare and that subfunctionalization better explains the observed distribution. More recently, non-toxic 3FP proteins have been found to be widely expressed in many different tissues in snakes, prompting the alternative hypothesis that proteins of restricted expression in saliva were selectively recruited for toxic functionality.
- Fry BG (March 2005). “From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins”. Genome Research. 15 (3): 403–20. doi:10.1101/gr.3228405. PMC 551567. PMID 15741511.
- Fry BG, Casewell NR, Wüster W, Vidal N, Young B, Jackson TN (September 2012). “The structural and functional diversification of the Toxicofera reptile venom system”. Toxicon. Advancing in Basic and Translational Venomics. 60 (4): 434–48. doi:10.1016/j.toxicon.2012.02.013. PMID 22446061.
- Reyes-Velasco J, Card DC, Andrew AL, Shaney KJ, Adams RH, Schield DR, Casewell NR, Mackessy SP, Castoe TA (January 2015). “Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom”. Molecular Biology and Evolution. 32 (1): 173–83. doi:10.1093/molbev/msu294. PMID 25338510.
- Hargreaves AD, Swain MT, Hegarty MJ, Logan DW, Mulley JF (August 2014). “Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins”. Genome Biology and Evolution. 6 (8): 2088–95. doi:10.1093/gbe/evu166. PMC 4231632. PMID 25079342.
References
- Nastopoulos V, Kanellopoulos PN, Tsernoglou D (September 1998). “Structure of dimeric and monomeric erabutoxin a refined at 1.5 A resolution” (PDF). Acta Crystallographica. Section D, Biological Crystallography. 54 (Pt 5): 964–74. Bibcode:1998AcCrD..54..964N. doi:10.1107/S0907444998005125. PMID 9757111.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Hegde RP, Rajagopalan N, Doley R, Kini M (2010). “Snake venom three-finger toxins”. In Mackessy SP (ed.). Handbook of venoms and toxins of reptiles. Boca Raton: CRC Press. pp. 287–302. ISBN 9781420008661.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- Utkin Y, Sunagar K, Jackson TN, Reeks T, Fry BG (2015). “Chapter 8: Three-finger toxins”. In Fry B (ed.). Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. Oxford University Press. pp. 218–227. ISBN 9780199309405.
- Sanz L, Pla D, Pérez A, Rodríguez Y, Zavaleta A, Salas M, Lomonte B, Calvete JJ (June 2016). “Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA₂ Dichotomy across Micrurus Venoms”. Toxins. 8 (6): 178. doi:10.3390/toxins8060178. PMC 4926144. PMID 27338473.
- Leath KJ, Johnson S, Roversi P, Hughes TR, Smith RA, Mackenzie L, Morgan BP, Lea SM (August 2007). “High-resolution structures of bacterially expressed soluble human CD59”. Acta Crystallographica. Section F, Structural Biology and Crystallization Communications. 63 (Pt 8): 648–52. doi:10.1107/S1744309107033477. PMC 2335151. PMID 17671359.
- Loughner CL, Bruford EA, McAndrews MS, Delp EE, Swamynathan S, Swamynathan SK (April 2016). “Organization, evolution and functions of the human and mouse Ly6/uPAR family genes”. Human Genomics. 10: 10. doi:10.1186/s40246-016-0074-2. PMC 4839075. PMID 27098205.
- Ploug M, Ellis V (August 1994). “Structure-function relationships in the receptor for urokinase-type plasminogen activator. Comparison to other members of the Ly-6 family and snake venom alpha-neurotoxins”. FEBS Letters. 349 (2): 163–8. doi:10.1016/0014-5793(94)00674-1. PMID 8050560. S2CID 86302713.
- Greenwald J, Fischer WH, Vale WW, Choe S (January 1999). “Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase”. Nature Structural Biology. 6 (1): 18–22. doi:10.1038/4887. PMID 9886286. S2CID 26301441.
- Kirsch T, Sebald W, Dreyer MK (June 2000). “Crystal structure of the BMP-2-BRIA ectodomain complex”. Nature Structural Biology. 7 (6): 492–6. doi:10.1038/75903. PMID 10881198. S2CID 19403233.
- Galat A (November 2008). “The three-fingered protein domain of the human genome”. Cellular and Molecular Life Sciences. 65 (21): 3481–93. doi:10.1007/s00018-008-8473-8. PMID 18821057. S2CID 19931506.
- Tsetlin VI (February 2015). “Three-finger snake neurotoxins and Ly6 proteins targeting nicotinic acetylcholine receptors: pharmacological tools and endogenous modulators”. Trends in Pharmacological Sciences. 36 (2): 109–23. doi:10.1016/j.tips.2014.11.003. PMID 25528970.
- Fry BG (March 2005). “From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins”. Genome Research. 15 (3): 403–20. doi:10.1101/gr.3228405. PMC 551567. PMID 15741511.
- Fry BG, Casewell NR, Wüster W, Vidal N, Young B, Jackson TN (September 2012). “The structural and functional diversification of the Toxicofera reptile venom system”. Toxicon. Advancing in Basic and Translational Venomics. 60 (4): 434–48. doi:10.1016/j.toxicon.2012.02.013. PMID 22446061.
- Reyes-Velasco J, Card DC, Andrew AL, Shaney KJ, Adams RH, Schield DR, Casewell NR, Mackessy SP, Castoe TA (January 2015). “Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom”. Molecular Biology and Evolution. 32 (1): 173–83. doi:10.1093/molbev/msu294. PMID 25338510.
- Hargreaves AD, Swain MT, Hegarty MJ, Logan DW, Mulley JF (August 2014). “Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins”. Genome Biology and Evolution. 6 (8): 2088–95. doi:10.1093/gbe/evu166. PMC 4231632. PMID 25079342.
- Nastopoulos V, Kanellopoulos PN, Tsernoglou D (September 1998). “Structure of dimeric and monomeric erabutoxin a refined at 1.5 A resolution”. Acta Crystallographica Section D. 54 (Pt 5): 964–74. doi:10.1107/S0907444998005125. PMID 9757111.
- Kini RM, Doley R (November 2010). “Structure, function and evolution of three-finger toxins: mini proteins with multiple targets”. Toxicon. 56 (6): 855–67. doi:10.1016/j.toxicon.2010.07.010. PMID 20670641.
- Hegde RP, Rajagopalan N, Doley R, Kini M (2010). “Snake venom three-finger toxins”. In Mackessy SP (ed.). Handbook of venoms and toxins of reptiles. Boca Raton: CRC Press. pp. 287–302. ISBN 9781420008661.
- Kessler P, Marchot P, Silva M, Servent D (August 2017). “The three-finger toxin fold: a multifunctional structural scaffold able to modulate cholinergic functions”. Journal of Neurochemistry. 142 (Suppl 2): 7–18. doi:10.1111/jnc.13975. PMID 28326549.
- Utkin Y, Sunagar K, Jackson T, Reeks T, Fry BG (2015). “Chapter 8: Three-finger toxins”. In Fry B (ed.). Venomous Reptiles and Their Toxins: Evolution, Pathophysiology and Biodiscovery. Oxford University Press. pp. 218–227. ISBN 9780199309405.
- “Snake three-finger toxin family”. VenomZone. Retrieved 21 April 2017.
- Dewan JC, Grant GA, Sacchettini JC (November 1994). “Crystal structure of kappa-bungarotoxin at 2.3-A resolution”. Biochemistry. 33 (44): 13147–54. doi:10.1021/bi00248a026. PMID 7947721.
- Pawlak J, Mackessy SP, Sixberry NM, Stura EA, Le Du MH, Ménez R, Foo CS, Ménez A, Nirthanan S, Kini RM (February 2009). “Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity”. FASEB Journal. 23 (2): 534–45. doi:10.1096/fj.08-113555. PMID 18952712. S2CID 4816592.
- Roy A, Zhou X, Chong MZ, D’hoedt D, Foo CS, Rajagopalan N, Nirthanan S, Bertrand D, Sivaraman J, Kini RM (March 2010). “Structural and functional characterization of a novel homodimeric three-finger neurotoxin from the venom of Ophiophagus hannah (king cobra)”. The Journal of Biological Chemistry. 285 (11): 8302–15. doi:10.1074/jbc.M109.074161. PMC 2832981. PMID 20071329.
- Osipov AV, Kasheverov IE, Makarova YV, Starkov VG, Vorontsova OV, Ziganshin RK, Andreeva TV, Serebryakova MV, Benoit A, Hogg RC, Bertrand D, Tsetlin VI, Utkin YN (May 2008). “Naturally occurring disulfide-bound dimers of three-fingered toxins: a paradigm for biological activity diversification”. The Journal of Biological Chemistry. 283 (21): 14571–80. doi:10.1074/jbc.M802085200. PMID 18381281.
- de la Rosa, Guillermo; Olvera, Felipe; Archundia, Irving G.; Lomonte, Bruno; Alagón, Alejandro; Corzo, Gerardo (December 2019). “Horse immunization with short-chain consensus α-neurotoxin generates antibodies against broad spectrum of elapid venomous species”. Nature Communications. 10 (1): 3642. Bibcode:2019NatCo..10.3642D. doi:10.1038/s41467-019-11639-2. ISSN 2041-1723. PMC 6692343. PMID 31409779.
- Nirthanan S, Gwee MC (January 2004). “Three-finger α-neurotoxins and the nicotinic acetylcholine receptor, forty years on”. Journal of Pharmacological Sciences. 94 (1): 1–17. doi:10.1254/jphs.94.1. PMID 14745112.
- Hassan-Puttaswamy V, Adams DJ, Kini RM (December 2015). “A Distinct Functional Site in Ω-Neurotoxins: Novel Antagonists of Nicotinic Acetylcholine Receptors from Snake Venom”. ACS Chemical Biology. 10 (12): 2805–15. doi:10.1021/acschembio.5b00492. PMID 26448325.
- Karlsson E, Mbugua PM, Rodriguez-Ithurralde D (1984-01-01). “Fasciculins, anticholinesterase toxins from the venom of the green mamba Dendroaspis angusticeps”. Journal de Physiologie. 79 (4): 232–40. PMID 6530667.
- Rajagopalan N, Pung YF, Zhu YZ, Wong PT, Kumar PP, Kini RM (November 2007). “Beta-cardiotoxin: a new three-finger toxin from Ophiophagus hannah (king cobra) venom with beta-blocker activity”. FASEB Journal. 21 (13): 3685–95. doi:10.1096/fj.07-8658com. PMID 17616557. S2CID 21235585.
- Diochot S, Baron A, Salinas M, Douguet D, Scarzello S, Dabert-Gay AS, Debayle D, Friend V, Alloui A, Lazdunski M, Lingueglia E (October 2012). “Black mamba venom peptides target acid-sensing ion channels to abolish pain” (PDF). Nature. 490 (7421): 552–5. Bibcode:2012Natur.490..552D. doi:10.1038/nature11494. PMID 23034652. S2CID 4337253.
- Fry BG, Wüster W, Kini RM, Brusic V, Khan A, Venkataraman D, Rooney AP (July 2003). “Molecular evolution and phylogeny of elapid snake venom three-finger toxins”. Journal of Molecular Evolution. 57 (1): 110–29. Bibcode:2003JMolE..57..110F. CiteSeerX 10.1.1.539.324. doi:10.1007/s00239-003-2461-2. PMID 12962311. S2CID 12358977.
- Reyes-Velasco J, Card DC, Andrew AL, Shaney KJ, Adams RH, Schield DR, Casewell NR, Mackessy SP, Castoe TA (January 2015). “Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom”. Molecular Biology and Evolution. 32 (1): 173–83. doi:10.1093/molbev/msu294. PMID 25338510.
- Zouridakis M, Giastas P, Zarkadas E, Chroni-Tzartou D, Bregestovski P, Tzartos SJ (November 2014). “Crystal structures of free and antagonist-bound states of human α9 nicotinic receptor extracellular domain”. Nature Structural & Molecular Biology. 21 (11): 976–80. doi:10.1038/nsmb.2900. PMID 25282151. S2CID 30096256.
- Trémeau O, Lemaire C, Drevet P, Pinkasfeld S, Ducancel F, Boulain JC, Ménez A (April 1995). “Genetic engineering of snake toxins. The functional site of Erabutoxin a, as delineated by site-directed mutagenesis, includes variant residues”. The Journal of Biological Chemistry. 270 (16): 9362–9. doi:10.1074/jbc.270.16.9362. PMID 7721859.
- Lu X, Davies J, Lu D, Xia M, Wattam B, Shang D, Sun Y, Scully M, Kakkar V (2006-01-01). “The effect of the single substitution of arginine within the RGD tripeptide motif of a modified neurotoxin dendroaspin on its activity of platelet aggregation and cell adhesion”. Cell Communication & Adhesion. 13 (3): 171–83. doi:10.1080/15419060600726183. PMID 16798616.
- Konshina AG, Boldyrev IA, Utkin YN, Omel’kov AV, Efremov RG (April 2011). “Snake cytotoxins bind to membranes via interactions with phosphatidylserine head groups of lipids”. PLOS ONE. 6 (4): e19064. Bibcode:2011PLoSO…619064K. doi:10.1371/journal.pone.0019064. PMC 3084733. PMID 21559494.
- Lee SC, Lin CC, Wang CH, Wu PL, Huang HW, Chang CI, Wu WG (July 2014). “Endocytotic routes of cobra cardiotoxins depend on spatial distribution of positively charged and hydrophobic domains to target distinct types of sulfated glycoconjugates on cell surface”. The Journal of Biological Chemistry. 289 (29): 20170–81. doi:10.1074/jbc.M114.557157. PMC 4106332. PMID 24898246.
- Fruchart-Gaillard C, Mourier G, Blanchet G, Vera L, Gilles N, Ménez R, Marcon E, Stura EA, Servent D (2012-06-14). “Engineering of three-finger fold toxins creates ligands with original pharmacological profiles for muscarinic and adrenergic receptors”. PLOS ONE. 7 (6): e39166. Bibcode:2012PLoSO…739166F. doi:10.1371/journal.pone.0039166. PMC 3375269. PMID 22720062.
- Georgieva D, Arni RK, Betzel C (December 2008). “Proteome analysis of snake venom toxins: pharmacological insights”. Expert Review of Proteomics. 5 (6): 787–97. doi:10.1586/14789450.5.6.787. PMID 19086859. S2CID 29126350.
- Saviola AJ, Peichoto ME, Mackessy SP (2014-12-01). “Rear-fanged snake venoms: an untapped source of novel compounds and potential drug leads”. Toxin Reviews. 33 (4): 185–201. doi:10.3109/15569543.2014.942040. hdl:11336/17030. ISSN 1556-9543. S2CID 30100277.
- Strydom, D. J. (December 1973). “Snake Venom Toxins: The Evolution of Some of the Toxins Found in Snake Venoms”. Systematic Zoology. 22 (4): 596–608. doi:10.2307/2412964. JSTOR 2412964.
- Fry BG (March 2005). “From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins”. Genome Research. 15 (3): 403–20. doi:10.1101/gr.3228405. PMC 551567. PMID 15741511.
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG (April 2013). “Complex cocktails: the evolutionary novelty of venoms”. Trends in Ecology & Evolution. 28 (4): 219–29. doi:10.1016/j.tree.2012.10.020. PMID 23219381.
- de la Rosa, Guillermo; Corrales-García, Ligia L.; Rodriguez-Ruiz, Ximena; López-Vera, Estuardo; Corzo, Gerardo (July 2018). “Short-chain consensus alpha-neurotoxin: a synthetic 60-mer peptide with generic traits and enhanced immunogenic properties”. Amino Acids. 50 (7): 885–895. doi:10.1007/s00726-018-2556-0. ISSN 0939-4451. PMID 29626299. S2CID 4638613.
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJ, Kerkkamp HM, Vos RA, Guerreiro I, Calvete JJ, Wüster W, Woods AE, Logan JM, Harrison RA, Castoe TA, de Koning AP, Pollock DD, Yandell M, Calderon D, Renjifo C, Currier RB, Salgado D, Pla D, Sanz L, Hyder AS, Ribeiro JM, Arntzen JW, van den Thillart GE, Boetzer M, Pirovano W, Dirks RP, Spaink HP, Duboule D, McGlinn E, Kini RM, Richardson MK (December 2013). “The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system”. Proceedings of the National Academy of Sciences of the United States of America. 110 (51): 20651–6. Bibcode:2013PNAS..11020651V. doi:10.1073/pnas.1314702110. PMC 3870661. PMID 24297900.
- Lauridsen LP, Laustsen AH, Lomonte B, Gutiérrez JM (March 2016). “Toxicovenomics and antivenom profiling of the Eastern green mamba snake (Dendroaspis angusticeps)” (PDF). Journal of Proteomics. 136: 248–61. doi:10.1016/j.jprot.2016.02.003. PMID 26877184.
- Sanz L, Pla D, Pérez A, Rodríguez Y, Zavaleta A, Salas M, Lomonte B, Calvete JJ (June 2016). “Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA₂ Dichotomy across Micrurus Venoms”. Toxins. 8 (6): 178. doi:10.3390/toxins8060178. PMC 4926144. PMID 27338473.
- Fry BG, Casewell NR, Wüster W, Vidal N, Young B, Jackson TN (September 2012). “The structural and functional diversification of the Toxicofera reptile venom system”. Toxicon. Advancing in Basic and Translational Venomics. 60 (4): 434–48. doi:10.1016/j.toxicon.2012.02.013. PMID 22446061.
- Hargreaves AD, Swain MT, Hegarty MJ, Logan DW, Mulley JF (August 2014). “Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins”. Genome Biology and Evolution. 6 (8): 2088–95. doi:10.1093/gbe/evu166. PMC 4231632. PMID 25079342.
- Sunagar K, Jackson TN, Undheim EA, Ali SA, Antunes A, Fry BG (November 2013). “Three-fingered RAVERs: Rapid Accumulation of Variations in Exposed Residues of snake venom toxins”. Toxins. 5 (11): 2172–208. doi:10.3390/toxins5112172. PMC 3847720. PMID 24253238.
External links
- SCOP: SSF57302
- CATH: 2.10.60.10


Leave a Reply