DREAM complex
The dimerization partner, RB-like, E2F and multi-vulval class B (DREAM) complex is a protein complex responsible for the regulation of cell cycle-dependent gene expression. The complex is evolutionarily conserved, although some of its components vary from species to species. In humans, the key proteins in the complex are RBL1 (p107) and RBL2 (p130), both of which are homologs of RB (p105) and bind repressive E2F transcription factors E2F4 and E2F5; DP1, DP2 and DP3, dimerization partners of E2F; and MuvB, which is a complex of LIN9/37/52/54 and RBBP4.
- Sadasivam, Subhashini; DeCaprio, James A. (11 July 2013). “The DREAM complex: master coordinator of cell cycle-dependent gene expression”. Nature Reviews Cancer. 13 (8): 585–595. doi:10.1038/nrc3556. PMC 3986830. PMID 23842645.
- Fischer, M; Müller, GA (December 2017). “Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes”. Critical Reviews in Biochemistry and Molecular Biology. 52 (6): 638–662. doi:10.1080/10409238.2017.1360836. PMID 28799433. S2CID 205695213.
Discovery
Genes encoding the MuvB complex were originally identified from loss-of-function mutation studies in C. elegans. When mutated, these genes produced worms with multiple vulva-like organs, hence the name ‘Muv’. Three classes of Muv genes were classified, with class B genes encoding homologues of mammalian RB, E2F, and DP1, and others such as LIN-54, LIN-37, LIN-7 and LIN-52, whose functions were not yet understood.
- Beitel, GJ; Lambie, EJ; Horvitz, HR (2000-08-22). “The C. elegans gene lin-9,which acts in an Rb-related pathway, is required for gonadal sheath cell development and encodes a novel protein”. Gene. 254 (1–2): 253–63. doi:10.1016/s0378-1119(00)00296-1. PMID 10974557.
- Thomas, JH; Ceol, CJ; Schwartz, HT; Horvitz, HR (May 2003). “New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans”. Genetics. 164 (1): 135–51. doi:10.1093/genetics/164.1.135. PMC 1462563. PMID 12750327.
Studies in Drosophila melanogaster ovarian follicle cells identified a protein complex that bound to repeatedly amplifying chorion genes. The complex included genes that had close homology with the MuvB genes such as Mip130, Mip120 and Mip40. These Mip genes were identified as homologues of the MuvB genes LIN9, LIN54, and LIN37 respectively. Further studies in the fly embryo nuclear extracts confirmed the coexistence of these proteins with others such as the RB homologues Rbf1 and Rbf2, and others like E2f and Dp. The protein complex was thus termed as the Drosophila RBF, E2f2 and Mip (dREAM) complex. Disruption of the dREAM complex through RNAi knockdown of the components of dREAM complex led to higher expression of E2f regulated genes that are typically silenced, implicating dREAM’s role in gene down-regulation. Later in Drosophila melanogaster, there was also found a testis-specific paralog of the Myb-MuvB/DREAM complex known as tMAC (testis-specific meiotic arrest complex), which is involved in meiotic arrest.
- Beall, EL; Manak, JR; Zhou, S; Bell, M; Lipsick, JS; Botchan, MR (2002-12-19). “Role for a Drosophila Myb-containing protein complex in site-specific DNA replication”. Nature. 420 (6917): 833–7. Bibcode:2002Natur.420..833B. doi:10.1038/nature01228. PMID 12490953. S2CID 4425307.
- Korenjak, M; Taylor-Harding, B; Binné, UK; Satterlee, JS; Stevaux, O; Aasland, R; White-Cooper, H; Dyson, N; Brehm, A (2004-10-15). “Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes”. Cell. 119 (2): 181–93. doi:10.1016/j.cell.2004.09.034. PMID 15479636. S2CID 17989678.
- Beall, E. L.; Lewis, P. W.; Bell, M.; Rocha, M.; Jones, D. L.; Botchan, M. R. (15 April 2007). “Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B”. Genes & Development. 21 (8): 904–919. doi:10.1101/gad.1516607. PMC 1847709. PMID 17403774.
A protein complex similar to dREAM was subsequently identified in C. elegans extract containing DP, RB, and MuvB, and was named as DRM. This complex included mammalian homologues of RB and DP, and other members of the MuvB complex.
- Harrison, MM; Ceol, CJ; Lu, X; Horvitz, HR (2006-11-07). “Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex”. Proceedings of the National Academy of Sciences of the United States of America. 103 (45): 16782–7. Bibcode:2006PNAS..10316782H. doi:10.1073/pnas.0608461103. PMC 1636532. PMID 17075059.
The mammalian DREAM complex was identified following immunoprecipitation of p130 with mass-spectrometry analysis. The results showed that p130 was associated with E2F4, E2F5, the dimerization partner DP, and LIN9, LIN54, LIN37, LIN52, and RBBP4 that make up the MuvB complex. Immunoprecipitation of MuvB factors also revealed association of BMYB. Subsequent immunoprecipitation with BMYB yielded all the MuvB core proteins, but not other members of the DREAM complex – p130, p107, E2F4/5 and DP. This indicated that MuvB associated with BMYB to form the BMYB-MuvB complex or with p130/p107, E2F4/5 and DP to form the DREAM complex. The DREAM complex was found prevalent in quiescent or starved cells, and the BMYB-MuvB complex was found in actively dividing cells, hinting at separate functionalities of these two complexes.
- Litovchick, L; Sadasivam, S; Florens, L; Zhu, X; Swanson, SK; Velmurugan, S; Chen, R; Washburn, MP; Liu, XS; DeCaprio, JA (2007-05-25). “Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence”. Molecular Cell. 26 (4): 539–51. doi:10.1016/j.molcel.2007.04.015. PMID 17531812.
MuvB-like complexes were also recently discovered in Arabidoposis that include E2F and MYB orthologs combined with LIN9 and LIN54 orthologs.
- Kobayashi, K; Suzuki, T; Iwata, E; Nakamichi, N; Suzuki, T; Chen, P; Ohtani, M; Ishida, T; Hosoya, H; Müller, S; Leviczky, T; Pettkó-Szandtner, A; Darula, Z; Iwamoto, A; Nomoto, M; Tada, Y; Higashiyama, T; Demura, T; Doonan, JH; Hauser, MT; Sugimoto, K; Umeda, M; Magyar, Z; Bögre, L; Ito, M (2015-08-04). “Transcriptional repression by MYB3R proteins regulates plant organ growth”. The EMBO Journal. 34 (15): 1992–2007. doi:10.15252/embj.201490899. PMC 4551348. PMID 26069325.
- Lang, Lucas; Pettkó-Szandtner, Aladár; Tunçay Elbaşı, Hasibe; Takatsuka, Hirotomo; Nomoto, Yuji; Zaki, Ahmad; Dorokhov, Stefan; De Jaeger, Geert; Eeckhout, Dominique; Ito, Masaki; Magyar, Zoltán; Bögre, László; Heese, Maren; Schnittger, Arp (December 2021). “The DREAM complex represses growth in response to DNA damage in Arabidopsis”. Life Science Alliance. 4 (12): e202101141. doi:10.26508/lsa.202101141. PMC 8500230. PMID 34583930.
Function
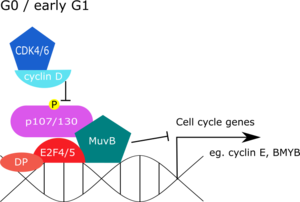
The main function of the DREAM complex is to repress G1/S and G2/M gene expression during quiescence (G0). Entry into the cell cycle dissociates p130 from the complex and leads to subsequent recruitment of activating E2F proteins. This allows for the expression of E2F regulated late G1 and S phase genes. BMYB (MYBL2), which is repressed by the DREAM complex during G0 is also able to be expressed at this time, and binds to MuvB during S phase to promote the expression of key G2/M phase genes such as CDK1 and CCNB1. FOXM1 is then recruited in G2 to further promote gene expression (e.g. AURKA). During late S phase BMYB is degraded via CUL1 (SCF complex), while FOXM1 is degraded during mitosis by the APC/C. Near the end of the cell cycle, the DREAM complex is re-assembled by DYRK1A to repress G1/S and G2/M genes.
- Sadasivam, S.; Duan, S.; DeCaprio, J. A. (5 March 2012). “The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression”. Genes & Development. 26 (5): 474–489. doi:10.1101/gad.181933.111. PMC 3305985. PMID 22391450.
- Sadasivam, Subhashini; DeCaprio, James A. (11 July 2013). “The DREAM complex: master coordinator of cell cycle-dependent gene expression”. Nature Reviews Cancer. 13 (8): 585–595. doi:10.1038/nrc3556. PMC 3986830. PMID 23842645.
The G1/S transition is a stage in the cell cycle at the boundary between the G1 phase, in which the cell grows, and the S phase, during which DNA is replicated. It is governed by cell cycle checkpoints to ensure cell cycle integrity and the subsequent S phase can pause in response to improperly or partially replicated DNA. During this transition the cell makes decisions to become quiescent (enter G0), differentiate, make DNA repairs, or proliferate based on environmental cues and molecular signaling inputs. The G1/S transition occurs late in G1 and the absence or improper application of this highly regulated checkpoint can lead to cellular transformation and disease states such as cancer. During this transition, G1 cyclin D-Cdk4/6 dimer phosphorylates retinoblastoma releasing transcription factor E2F, which then drives the transition from G1 to S phase. The G1/S transition is highly regulated by transcription factor p53 in order to halt the cell cycle when DNA is damaged. It is a “point of no return” beyond which the cell is committed to dividing; in yeast this is called the Start point, and in multicellular eukaryotes it is termed the restriction point (R-Point). If a cell passes through the G1/S transition the cell will continue through the cell cycle regardless of incoming mitogenic factors due to the positive feed-back loop of G1-S transcription. Positive feed-back loops include G1 cyclins and accumulation of E2F.
- Bartek J, Lukas J (February 2001). “Pathways governing G1/S transition and their response to DNA damage”. FEBS Letters. 490 (3): 117–22. doi:10.1016/S0014-5793(01)02114-7. PMID 11223026. S2CID 16090531.
- Bertoli C, Skotheim JM, de Bruin RA (August 2013). “Control of cell cycle transcription during G1 and S phases”. Nature Reviews Molecular Cell Biology. 14 (8): 518–28. doi:10.1038/nrm3629. PMC 4569015. PMID 23877564.
- Massagué J (November 2004). “G1 cell-cycle control and cancer”. Nature. 432 (7015): 298–306. Bibcode:2004Natur.432..298M. doi:10.1038/nature03094. PMID 15549091. S2CID 4428026.
- Bartek J, Lukas J (February 2001). “Pathways governing G1/S transition and their response to DNA damage”. FEBS Letters. 490 (3): 117–22. doi:10.1016/S0014-5793(01)02114-7. PMID 11223026. S2CID 16090531.
- Lodish H, Berk A, Kaiser C, Krieger M (2012). Molecular Cell Biology (7th ed.). Freeman, W. H. & Company. ISBN 978-1-4641-0981-2.
- Tenga MJ, Lazar IM (January 2013). “Proteomic snapshot of breast cancer cell cycle: G1/S transition point”. Proteomics. 13 (1): 48–60. doi:10.1002/pmic.201200188. PMC 4123745. PMID 23152136.
The G2-M DNA damage checkpoint is an important cell cycle checkpoint in eukaryotic organisms that ensures that cells don’t initiate mitosis until damaged or incompletely replicated DNA is sufficiently repaired. Cells with a defective G2-M checkpoint will undergo apoptosis or death after cell division if they enter the M phase before repairing their DNA. The defining biochemical feature of this checkpoint is the activation of M-phasecyclin-CDK complexes, which phosphorylate proteins that promote spindle assembly and bring the cell to metaphase. The cell cycle is driven by proteins called cyclin dependent kinases that associate with cyclin regulatory proteins at different checkpoints of the cell cycle. Different phases of the cell cycle experience activation and/or deactivation of specific cyclin-CDK complexes. CyclinB-CDK1 activity is specific to the G2/M checkpoint. Accumulation of cyclin B increases the activity of the cyclin dependent kinase Cdk1 human homolog Cdc2 as cells prepare to enter mitosis. Cdc2 activity is further regulated by phosphorylation/dephosphorylation of its corresponding activators and inhibitors. Through a positive feedback loop, CyclinB-Cdc2 activates the phosphatase Cdc25 which in turn deactivates the CyclinB-Cdc2 inhibitors, Wee1 and Myt1. Cdc25 activates the complex through the removal of phosphates from the active site while Wee1 inactivates the complex through the phosphorylation of tyrosine residues, specifically tyrosine-15. This loop is further amplified indirectly through the coordinated interaction of the Aurora A kinase and the Bora cofactor. During the G2 phase, Bora accumulates and forms an activation complex with Aurora A. This complex then regulates the activation of Polo-like kinase 1 (Plk1). Plk1 phosphorylates Wee1, targeting it for degradation through the SCF ubiquitin ligase complex (SCF complex), and activates Cdc25 through phosphorylation with combined action activating Cdc2. The combined activity and complex of Cdc2, Cdc25, and Plk1 with the accumulation of cyclin B activates the CyclinB-Cdc2 complex, promoting entry into mitosis. Many proteins involved in this positive feedback loop drive the activation of the CyclinB-Cdc2 complex because entry into mitosis requires an all-or-none response. The Novak-Tyson model is a mathematical model used to explain such regulatory loop that predicted the irreversible transition into mitosis driven by hysteresis. Through experiments in Xenopus laevis cell-free egg extracts, such model was confirmed as the basis for entry into mitosis. Once cyclin concentration reaches a certain minimum activation threshold, Cdc2 is rapidly activated. It remains in this state until activity falls below a separate inactivation threshold at which it is abruptly inactivated through tyrosine phosphorylation by Wee1 and Myt1. In the case of unreplicated DNA, the cyclin concentration threshold for Cdc2 activation is further increased. Through this mechanism, there exists two separate steady-state conditions separated by an unstable steady state. The bistable and hysteretic nature of CyclinB-Cdc2 ensures a highly regulated nature of the G2/M checkpoint.
- Cuddihy, Andrew R.; O’Connell, Matthew J. (2003). “Cell-cycle responses to DNA damage in G2”. International Review of Cytology. 222: 99–140. doi:10.1016/s0074-7696(02)22013-6. ISBN 9780123646262. ISSN 0074-7696. PMID 12503848.
- Morgan, David Owen, 1958- (2007). The cell cycle : principles of control. London: New Science Press. ISBN 978-0-19-920610-0. OCLC 70173205.
- Gould, K. L.; Nurse, P. (1989). “Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis”. Nature. 342 (6245): 39–45. Bibcode:1989Natur.342…39G. doi:10.1038/342039a0. PMID 2682257. S2CID 4287870.
- Seki, A.; Coppinger, J. A.; Jang, C.-Y.; Yates, J. R.; Fang, G. (20 June 2008). “Bora and the Kinase Aurora A Cooperatively Activate the Kinase Plk1 and Control Mitotic Entry”. Science. 320 (5883): 1655–1658. Bibcode:2008Sci…320.1655S. doi:10.1126/science.1157425. PMC 2834883. PMID 18566290.
- Novak, B.; Tyson, J. J. (1993). “Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos”. Journal of Cell Science. 106 (4): 1153–1168. doi:10.1242/jcs.106.4.1153. PMID 8126097.
- Sha, Wei; et al. (September 2002). “Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts”. Proceedings of the National Academy of Sciences. 100 (3): 975–980. Bibcode:2003PNAS..100..975S. doi:10.1073/pnas.0235349100. PMC 298711. PMID 12509509.
G0
During quiescence, the DREAM complex represses G1/S and G2/M gene expression. In mammalian systems, chromatin-immunoprecipitation (ChIP) studies have revealed that DREAM components are found together at promoters of genes that peak in G1/S or G2/M phase. Abrogation of the DREAM complex on the other hand, led to increased expression of E2F regulated genes normally repressed in the G0 phase. Contrary to mammalian cells, the fly dREAM complex was found at almost one-third of all promoters, which may reflect a broader role for dREAM in gene regulation, such as programmed cell death of neural precursor cells.
- Kobayashi, K; Suzuki, T; Iwata, E; Nakamichi, N; Suzuki, T; Chen, P; Ohtani, M; Ishida, T; Hosoya, H; Müller, S; Leviczky, T; Pettkó-Szandtner, A; Darula, Z; Iwamoto, A; Nomoto, M; Tada, Y; Higashiyama, T; Demura, T; Doonan, JH; Hauser, MT; Sugimoto, K; Umeda, M; Magyar, Z; Bögre, L; Ito, M (2015-08-04). “Transcriptional repression by MYB3R proteins regulates plant organ growth”. The EMBO Journal. 34 (15): 1992–2007. doi:10.15252/embj.201490899. PMC 4551348. PMID 26069325.
- Litovchick, L; Florens, LA; Swanson, SK; Washburn, MP; DeCaprio, JA (2011-04-15). “DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly”. Genes & Development. 25 (8): 801–13. doi:10.1101/gad.2034211. PMC 3078706. PMID 21498570.
- Georlette, D; Ahn, S; MacAlpine, DM; Cheung, E; Lewis, PW; Beall, EL; Bell, SP; Speed, T; Manak, JR; Botchan, MR (2007-11-15). “Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells”. Genes & Development. 21 (22): 2880–96. doi:10.1101/gad.1600107. PMC 2049191. PMID 17978103.
- Rovani, Margritte K.; Brachmann, Carrie Baker; Ramsay, Gary; Katzen, Alisa L. (December 2012). “The dREAM/Myb–MuvB complex and Grim are key regulators of the programmed death of neural precursor cells at the Drosophila posterior wing margin”. Developmental Biology. 372 (1): 88–102. doi:10.1016/j.ydbio.2012.08.022. PMC 3621911. PMID 22960039.
Docking of the DREAM complex to promoters is achieved by binding of LIN-54 to regions known as cell cycle genes homology region (CHR). These are specific sequence of nucleotides that are commonly found in the promoters of genes expressed during late S phase or G2/M phase. Docking can also be achieved via E2F proteins binding to sequences known as cell cycle-dependent element sites (CDEs). Some cell cycle dependent genes have been found where both CHRs and CDEs are in proximity to one another. Because p130-E2F4 can form stable associations with the MuvB complex, the proximity of CHRs to CDEs suggests that affinity of binding of the DREAM complex to target genes is cooperatively improved by association with both the binding sites.
- Müller, GA; Engeland, K (February 2010). “The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription”. The FEBS Journal. 277 (4): 877–93. doi:10.1111/j.1742-4658.2009.07508.x. PMID 20015071. S2CID 8955433.
When DREAM is docked onto the promoter, p130 is bound to LIN52, and this association inhibits LIN52 binding to chromatin modifier proteins. Therefore, unlike RB-E2F, the DREAM complex is unlikely to directly recruit chromatin modifiers to repress gene expression, although some associations have been suggested. DREAM complex may instead down-regulate gene expression by affecting nucleosome positioning. Compacted DNA at transcription start sites inhibit gene expression by blocking the docking of RNA polymerase. In worms for example, loss of a MuvB complex protein, LIN35, leads to loss of repressive histone associations and high expression of cell cycle dependent genes. However, direct evidence for the link between repressive histones and the DREAM complex remains to be elucidated.
- Forristal, C; Henley, SA; MacDonald, JI; Bush, JR; Ort, C; Passos, DT; Talluri, S; Ishak, CA; Thwaites, MJ; Norley, CJ; Litovchick, L; DeCaprio, JA; DiMattia, G; Holdsworth, DW; Beier, F; Dick, FA (June 2014). “Loss of the mammalian DREAM complex deregulates chondrocyte proliferation”. Molecular and Cellular Biology. 34 (12): 2221–34. doi:10.1128/MCB.01523-13. PMC 4054284. PMID 24710275.
- Guiley, KZ; Liban, TJ; Felthousen, JG; Ramanan, P; Litovchick, L; Rubin, SM (2015-05-01). “Structural mechanisms of DREAM complex assembly and regulation”. Genes & Development. 29 (9): 961–74. doi:10.1101/gad.257568.114. PMC 4421984. PMID 25917549.
- Sandoval, R; Pilkinton, M; Colamonici, OR (2009-10-15). “Deletion of the p107/p130-binding domain of Mip130/LIN-9 bypasses the requirement for CDK4 activity for the dissociation of Mip130/LIN-9 from p107/p130-E2F4 complex”. Experimental Cell Research. 315 (17): 2914–20. doi:10.1016/j.yexcr.2009.07.014. PMC 2757496. PMID 19619530.
- Stiegler, P; De Luca, A; Bagella, L; Giordano, A (1998-11-15). “The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter”. Cancer Research. 58 (22): 5049–52. PMID 9823308.
- Bai, L; Morozov, AV (November 2010). “Gene regulation by nucleosome positioning”. Trends in Genetics. 26 (11): 476–83. doi:10.1016/j.tig.2010.08.003. PMID 20832136.
- Latorre, I; Chesney, MA; Garrigues, JM; Stempor, P; Appert, A; Francesconi, M; Strome, S; Ahringer, J (2015-03-01). “The DREAM complex promotes gene body H2A.Z for target repression”. Genes & Development. 29 (5): 495–500. doi:10.1101/gad.255810.114. PMC 4358402. PMID 25737279.
G1/S
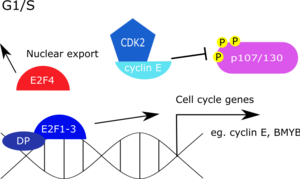
Like its counterpart, RB-E2F, the DREAM complex is also affected by similar growth stimuli and subsequent cyclin-CDK activity. Increasing cyclin D-CDK4 and cyclin E-CDK2 activity dissociates the DREAM complex from the promoter by phosphorylation of p130. Hyper-phosphorylated p130 is subsequently degraded and E2F4 exported from the nucleus. Once the repressive E2Fs are vacated, activating E2Fs bind to the promoter to up-regulate G1/S genes that promote DNA synthesis and transition of the cell cycle. BMYB is also up-regulated during this time, which then binds to genes that peak at G2/M phase. Binding of BMYB to late cell cycle genes is dependent on its association with the MuvB core to form the BMYB-MuvB complex, which is then able to up-regulate genes in the G2/M phase.
- Tedesco, D; Lukas, J; Reed, SI (2002-11-15). “The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2)”. Genes & Development. 16 (22): 2946–57. doi:10.1101/gad.1011202. PMC 187481. PMID 12435635.
- Bhattacharya, S; Garriga, J; Calbó, J; Yong, T; Haines, DS; Graña, X (2003-04-24). “SKP2 associates with p130 and accelerates p130 ubiquitylation and degradation in human cells”. Oncogene. 22 (16): 2443–51. doi:10.1038/sj.onc.1206339. PMID 12717421. S2CID 26125392.
- Gaubatz, S; Lees, JA; Lindeman, GJ; Livingston, DM (February 2001). “E2F4 is exported from the nucleus in a CRM1-dependent manner”. Molecular and Cellular Biology. 21 (4): 1384–92. doi:10.1128/MCB.21.4.1384-1392.2001. PMC 99590. PMID 11158323.
- Takahashi, Y; Rayman, JB; Dynlacht, BD (2000-04-01). “Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression”. Genes & Development. 14 (7): 804–16. doi:10.1101/gad.14.7.804. PMC 316494. PMID 10766737.
- Lam, EW; Robinson, C; Watson, RJ (September 1992). “Characterization and cell cycle-regulated expression of mouse B-myb”. Oncogene. 7 (9): 1885–90. PMID 1501895.
- Pilkinton, M; Sandoval, R; Song, J; Ness, SA; Colamonici, OR (2007-01-05). “Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1”. The Journal of Biological Chemistry. 282 (1): 168–75. doi:10.1074/jbc.M609924200. PMID 17098733. S2CID 21963932.
- Guiley, KZ; Liban, TJ; Felthousen, JG; Ramanan, P; Litovchick, L; Rubin, SM (2015-05-01). “Structural mechanisms of DREAM complex assembly and regulation”. Genes & Development. 29 (9): 961–74. doi:10.1101/gad.257568.114. PMC 4421984. PMID 25917549.
- Sadasivam, S.; Duan, S.; DeCaprio, J. A. (5 March 2012). “The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression”. Genes & Development. 26 (5): 474–489. doi:10.1101/gad.181933.111. PMC 3305985. PMID 22391450.
- Litovchick, L; Sadasivam, S; Florens, L; Zhu, X; Swanson, SK; Velmurugan, S; Chen, R; Washburn, MP; Liu, XS; DeCaprio, JA (2007-05-25). “Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence”. Molecular Cell. 26 (4): 539–51. doi:10.1016/j.molcel.2007.04.015. PMID 17531812.
Late mitosis
Near the end of mitosis, p130 and p107 are dephosphorylated from their hyperphosphorylated state by the phosphatase PP2a. Inhibition of PP2a activity reduced promoter binding of some of the proteins of the DREAM complex in the subsequent G1 phase and de-repression of gene expression.
- Kolupaeva, V; Janssens, V (January 2013). “PP1 and PP2A phosphatases–cooperating partners in modulating retinoblastoma protein activation”. The FEBS Journal. 280 (2): 627–43. doi:10.1111/j.1742-4658.2012.08511.x. PMID 22299668. S2CID 46705471.
- Kurimchak, A; Graña, X (2015). “PP2A: more than a reset switch to activate pRB proteins during the cell cycle and in response to signaling cues”. Cell Cycle. 14 (1): 18–30. doi:10.4161/15384101.2014.985069. PMC 4612414. PMID 25483052.
- Naetar, N; Soundarapandian, V; Litovchick, L; Goguen, KL; Sablina, AA; Bowman-Colin, C; Sicinski, P; Hahn, WC; DeCaprio, JA; Livingston, DM (2014-06-19). “PP2A-mediated regulation of Ras signaling in G2 is essential for stable quiescence and normal G1 length”. Molecular Cell. 54 (6): 932–45. doi:10.1016/j.molcel.2014.04.023. PMC 4118046. PMID 24857551.
Other components have been shown to be phosphorylated for DREAM complex assembly to occur. Of these, LIN52 phosphorylation on its S28 residue is the most well-understood. Substitution of this serine to alanine led to reduced binding of the MuvB core to p130 and impaired the ability of cells to enter quiescence. This indicates that LIN52 S28 phosphorylation is required for proper association and function of the DREAM complex via binding with p130. One known regulator of phosphorylation of the S28 residue is the DYRK1A. The loss of this kinase leads to decreased phosphorylation of the S28 residue and association of p130 with MuvB. DYRK1A was also found to degrade cyclin D1, which would increase p21 levels – both of which contribute to cell cycle exit.
- Chen, JY; Lin, JR; Tsai, FC; Meyer, T (2013-10-10). “Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle”. Molecular Cell. 52 (1): 87–100. doi:10.1016/j.molcel.2013.09.009. PMC 4039290. PMID 24119401.
- Litovchick, L; Florens, LA; Swanson, SK; Washburn, MP; DeCaprio, JA (2011-04-15). “DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly”. Genes & Development. 25 (8): 801–13. doi:10.1101/gad.2034211. PMC 3078706. PMID 21498570.
The DREAM complex was also shown to regulate cytokinesis through GAS2L3.
- Wolter, P; Schmitt, K; Fackler, M; Kremling, H; Probst, L; Hauser, S; Gruss, OJ; Gaubatz, S (15 May 2012). “GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability”. Journal of Cell Science. 125 (Pt 10): 2393–406. doi:10.1242/jcs.097253. PMID 22344256.
Cancer therapy
Due to its regulatory role in the cell cycle, targeting the DREAM complex might enhance anticancer treatments such as imatinib.
- DeCaprio, James A.; Duensing, Anette (July 2014). “The DREAM complex in antitumor activity of imatinib mesylate in gastrointestinal stromal tumors”. Current Opinion in Oncology. 26 (4): 415–421. doi:10.1097/CCO.0000000000000090. PMC 4236229. PMID 24840522.
- Boichuk, S.; Parry, J. A.; Makielski, K. R.; Litovchick, L.; Baron, J. L.; Zewe, J. P.; Wozniak, A.; Mehalek, K. R.; Korzeniewski, N.; Seneviratne, D. S.; Schoffski, P.; Debiec-Rychter, M.; DeCaprio, J. A.; Duensing, A. (20 June 2013). “The DREAM Complex Mediates GIST Cell Quiescence and Is a Novel Therapeutic Target to Enhance Imatinib-Induced Apoptosis”. Cancer Research. 73 (16): 5120–5129. doi:10.1158/0008-5472.CAN-13-0579. PMID 23786773.
See also
- Pocket protein family
- Pocket protein family consists of three proteins:
- RB – Retinoblastoma protein
- p107 – Retinoblastoma-like protein 1
- p130 – Retinoblastoma-like protein 2
- They play crucial roles in the metazoan cell cycle through interaction with members of the E2F transcription factors family.
- Cobrinik D (2005). “Pocket proteins and cell cycle control”. Oncogene. 24 (17): 2796–809. doi:10.1038/sj.onc.1208619. PMID 15838516.
- Pocket protein family consists of three proteins:
- Retinoblastoma-like 1 (p107), also known as RBL1, is a protein that in humans is encoded by the RBL1 gene. The protein encoded by this gene is similar in sequence and possibly function to the product of the retinoblastoma 1 (RB1) gene. The RB1 gene product is a tumor suppressor protein that appears to be involved in cell cycle regulation, as it is phosphorylated in the S to M phase transition and is dephosphorylated in the G1 phase of the cell cycle. Both the RB1 protein and the product of this gene can form a complex with adenovirus E1A protein and SV40 Large T-antigen, with the SV40 large T-antigen binding only to the unphosphorylated form of each protein. In addition, both proteins can inhibit the transcription of cell cycle genes containing E2F binding sites in their promoters. Due to the sequence and biochemical similarities with the RB1 protein, it is thought that the protein encoded by this gene may also be a tumor suppressor. Two transcript variants encoding different isoforms have been found for this gene. Retinoblastoma-like protein 1 has been shown to interact with:
- BEGAIN,
- BRCA1,
- BRF1,
- Cyclin A2,
- Cyclin-dependent kinase 2,
- E2F1,
- HDAC1,
- MYBL2
- Mothers against decapentaplegic homolog 3,
- Prohibitin, and
- RBBP8.
- GRCh38: Ensembl release 89: ENSG00000080839 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000027641 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Entrez Gene: RBL1 retinoblastoma-like 1 (p107)”.
- Ewen ME, Xing YG, Lawrence JB, Livingston DM (Sep 1991). “Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein”. Cell. 66 (6): 1155–64. doi:10.1016/0092-8674(91)90038-Z. PMID 1833063. S2CID 27478008.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct 2005). “Towards a proteome-scale map of the human protein-protein interaction network”. Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- Fan S, Yuan R, Ma YX, Xiong J, Meng Q, Erdos M, Zhao JN, Goldberg ID, Pestell RG, Rosen EM (Aug 2001). “Disruption of BRCA1 LXCXE motif alters BRCA1 functional activity and regulation of RB family but not RB protein binding”. Oncogene. 20 (35): 4827–41. doi:10.1038/sj.onc.1204666. PMID 11521194.
- Sutcliffe JE, Cairns CA, McLees A, Allison SJ, Tosh K, White RJ (Jun 1999). “RNA polymerase III transcription factor IIIB is a target for repression by pocket proteins p107 and p130”. Molecular and Cellular Biology. 19 (6): 4255–61. doi:10.1128/mcb.19.6.4255. PMC 104385. PMID 10330166.
- Dyson N, Dembski M, Fattaey A, Ngwu C, Ewen M, Helin K (Dec 1993). “Analysis of p107-associated proteins: p107 associates with a form of E2F that differs from pRB-associated E2F-1”. Journal of Virology. 67 (12): 7641–7. doi:10.1128/JVI.67.12.7641-7647.1993. PMC 238233. PMID 8230483.
- Joaquin M, Bessa M, Saville MK, Watson RJ (Nov 2002). “B-Myb overcomes a p107-mediated cell proliferation block by interacting with an N-terminal domain of p107”. Oncogene. 21 (52): 7923–32. doi:10.1038/sj.onc.1206001. PMID 12439743. S2CID 21761703.
- Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E (Feb 1999). “Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest”. Molecular and Cellular Biology. 19 (2): 1460–9. doi:10.1128/mcb.19.2.1460. PMC 116074. PMID 9891079.
- Leng X, Noble M, Adams PD, Qin J, Harper JW (Apr 2002). “Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4”. Molecular and Cellular Biology. 22 (7): 2242–54. doi:10.1128/mcb.22.7.2242-2254.2002. PMC 133692. PMID 11884610.
- Lai A, Lee JM, Yang WM, DeCaprio JA, Kaelin WG, Seto E, Branton PE (Oct 1999). “RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins”. Molecular and Cellular Biology. 19 (10): 6632–41. doi:10.1128/mcb.19.10.6632. PMC 84642. PMID 10490602.
- Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D (Sep 1998). “The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase”. Proceedings of the National Academy of Sciences of the United States of America. 95 (18): 10493–8. Bibcode:1998PNAS…9510493F. doi:10.1073/pnas.95.18.10493. PMC 27922. PMID 9724731.
- Joaquin M, Watson RJ (Nov 2003). “The cell cycle-regulated B-Myb transcription factor overcomes cyclin-dependent kinase inhibitory activity of p57(KIP2) by interacting with its cyclin-binding domain”. The Journal of Biological Chemistry. 278 (45): 44255–64. doi:10.1074/jbc.M308953200. PMID 12947099.
- Chen CR, Kang Y, Siegel PM, Massagué J (Jul 2002). “E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression”. Cell. 110 (1): 19–32. doi:10.1016/s0092-8674(02)00801-2. PMID 12150994. S2CID 8945574.
- Wang S, Nath N, Adlam M, Chellappan S (Jun 1999). “Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function”. Oncogene. 18 (23): 3501–10. doi:10.1038/sj.onc.1202684. PMID 10376528. S2CID 33828482.
- Fusco C, Reymond A, Zervos AS (Aug 1998). “Molecular cloning and characterization of a novel retinoblastoma-binding protein”. Genomics. 51 (3): 351–8. doi:10.1006/geno.1998.5368. PMID 9721205.
- Retinoblastoma-like protein 2 is a protein that in humans is encoded by the RBL2 gene. Retinoblastoma-like protein 2 has been shown to interact with:
- BRCA1,
- BRF1
- C-Raf,
- Cyclin E1,
- Cyclin-dependent kinase 2,
- HDAC1,
- Prohibitin, and
- RBBP8
- GRCh38: Ensembl release 89: ENSG00000103479 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000031666 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- Mayol X, Graña X, Baldi A, Sang N, Hu Q, Giordano A (Sep 1993). “Cloning of a new member of the retinoblastoma gene family (pRb2) which binds to the E1A transforming domain”. Oncogene. 8 (9): 2561–6. PMID 8361765.
- Baldi A, Boccia V, Claudio PP, De Luca A, Giordano A (May 1996). “Genomic structure of the human retinoblastoma-related Rb2/p130 gene”. Proceedings of the National Academy of Sciences of the United States of America. 93 (10): 4629–32. Bibcode:1996PNAS…93.4629B. doi:10.1073/pnas.93.10.4629. PMC 39329. PMID 8643454.
- Fan S, Yuan R, Ma YX, Xiong J, Meng Q, Erdos M, Zhao JN, Goldberg ID, Pestell RG, Rosen EM (Aug 2001). “Disruption of BRCA1 LXCXE motif alters BRCA1 functional activity and regulation of RB family but not RB protein binding”. Oncogene. 20 (35): 4827–41. doi:10.1038/sj.onc.1204666. PMID 11521194.
- Sutcliffe JE, Cairns CA, McLees A, Allison SJ, Tosh K, White RJ (Jun 1999). “RNA polymerase III transcription factor IIIB is a target for repression by pocket proteins p107 and p130”. Molecular and Cellular Biology. 19 (6): 4255–61. doi:10.1128/mcb.19.6.4255. PMC 104385. PMID 10330166.
- Wang S, Ghosh RN, Chellappan SP (Dec 1998). “Raf-1 physically interacts with Rb and regulates its function: a link between mitogenic signaling and cell cycle regulation”. Molecular and Cellular Biology. 18 (12): 7487–98. doi:10.1128/mcb.18.12.7487. PMC 109329. PMID 9819434.
- Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E (Feb 1999). “Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest”. Molecular and Cellular Biology. 19 (2): 1460–9. doi:10.1128/mcb.19.2.1460. PMC 116074. PMID 9891079.
- Li Y, Graham C, Lacy S, Duncan AM, Whyte P (Dec 1993). “The adenovirus E1A-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E”. Genes & Development. 7 (12A): 2366–77. doi:10.1101/gad.7.12a.2366. PMID 8253383.
- Lacy S, Whyte P (May 1997). “Identification of a p130 domain mediating interactions with cyclin A/cdk 2 and cyclin E/cdk 2 complexes”. Oncogene. 14 (20): 2395–406. doi:10.1038/sj.onc.1201085. PMID 9188854. S2CID 26359262.
- Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D (Sep 1998). “The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase”. Proceedings of the National Academy of Sciences of the United States of America. 95 (18): 10493–8. Bibcode:1998PNAS…9510493F. doi:10.1073/pnas.95.18.10493. PMC 27922. PMID 9724731.
- Bouzahzah B, Fu M, Iavarone A, Factor VM, Thorgeirsson SS, Pestell RG (Aug 2000). “Transforming growth factor-beta1 recruits histone deacetylase 1 to a p130 repressor complex in transgenic mice in vivo”. Cancer Research. 60 (16): 4531–7. PMID 10969803.
- Wang S, Nath N, Adlam M, Chellappan S (Jun 1999). “Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function”. Oncogene. 18 (23): 3501–10. doi:10.1038/sj.onc.1202684. PMID 10376528. S2CID 33828482.
- Meloni AR, Smith EJ, Nevins JR (Aug 1999). “A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor”. Proceedings of the National Academy of Sciences of the United States of America. 96 (17): 9574–9. Bibcode:1999PNAS…96.9574M. doi:10.1073/pnas.96.17.9574. PMC 22250. PMID 10449734.
- Fusco C, Reymond A, Zervos AS (Aug 1998). “Molecular cloning and characterization of a novel retinoblastoma-binding protein”. Genomics. 51 (3): 351–8. doi:10.1006/geno.1998.5368. PMID 9721205.
References
- Sadasivam, Subhashini; DeCaprio, James A. (11 July 2013). “The DREAM complex: master coordinator of cell cycle-dependent gene expression”. Nature Reviews Cancer. 13 (8): 585–595. doi:10.1038/nrc3556. PMC 3986830. PMID 23842645.
- Fischer, M; Müller, GA (December 2017). “Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes”. Critical Reviews in Biochemistry and Molecular Biology. 52 (6): 638–662. doi:10.1080/10409238.2017.1360836. PMID 28799433. S2CID 205695213.
- Beitel, GJ; Lambie, EJ; Horvitz, HR (2000-08-22). “The C. elegans gene lin-9,which acts in an Rb-related pathway, is required for gonadal sheath cell development and encodes a novel protein”. Gene. 254 (1–2): 253–63. doi:10.1016/s0378-1119(00)00296-1. PMID 10974557.
- Thomas, JH; Ceol, CJ; Schwartz, HT; Horvitz, HR (May 2003). “New genes that interact with lin-35 Rb to negatively regulate the let-60 ras pathway in Caenorhabditis elegans”. Genetics. 164 (1): 135–51. doi:10.1093/genetics/164.1.135. PMC 1462563. PMID 12750327.
- Beall, EL; Manak, JR; Zhou, S; Bell, M; Lipsick, JS; Botchan, MR (2002-12-19). “Role for a Drosophila Myb-containing protein complex in site-specific DNA replication”. Nature. 420 (6917): 833–7. Bibcode:2002Natur.420..833B. doi:10.1038/nature01228. PMID 12490953. S2CID 4425307.
- Korenjak, M; Taylor-Harding, B; Binné, UK; Satterlee, JS; Stevaux, O; Aasland, R; White-Cooper, H; Dyson, N; Brehm, A (2004-10-15). “Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes”. Cell. 119 (2): 181–93. doi:10.1016/j.cell.2004.09.034. PMID 15479636. S2CID 17989678.
- Beall, E. L.; Lewis, P. W.; Bell, M.; Rocha, M.; Jones, D. L.; Botchan, M. R. (15 April 2007). “Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B”. Genes & Development. 21 (8): 904–919. doi:10.1101/gad.1516607. PMC 1847709. PMID 17403774.
- Harrison, MM; Ceol, CJ; Lu, X; Horvitz, HR (2006-11-07). “Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex”. Proceedings of the National Academy of Sciences of the United States of America. 103 (45): 16782–7. Bibcode:2006PNAS..10316782H. doi:10.1073/pnas.0608461103. PMC 1636532. PMID 17075059.
- Litovchick, L; Sadasivam, S; Florens, L; Zhu, X; Swanson, SK; Velmurugan, S; Chen, R; Washburn, MP; Liu, XS; DeCaprio, JA (2007-05-25). “Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence”. Molecular Cell. 26 (4): 539–51. doi:10.1016/j.molcel.2007.04.015. PMID 17531812.
- Kobayashi, K; Suzuki, T; Iwata, E; Nakamichi, N; Suzuki, T; Chen, P; Ohtani, M; Ishida, T; Hosoya, H; Müller, S; Leviczky, T; Pettkó-Szandtner, A; Darula, Z; Iwamoto, A; Nomoto, M; Tada, Y; Higashiyama, T; Demura, T; Doonan, JH; Hauser, MT; Sugimoto, K; Umeda, M; Magyar, Z; Bögre, L; Ito, M (2015-08-04). “Transcriptional repression by MYB3R proteins regulates plant organ growth”. The EMBO Journal. 34 (15): 1992–2007. doi:10.15252/embj.201490899. PMC 4551348. PMID 26069325.
- Lang, Lucas; Pettkó-Szandtner, Aladár; Tunçay Elbaşı, Hasibe; Takatsuka, Hirotomo; Nomoto, Yuji; Zaki, Ahmad; Dorokhov, Stefan; De Jaeger, Geert; Eeckhout, Dominique; Ito, Masaki; Magyar, Zoltán; Bögre, László; Heese, Maren; Schnittger, Arp (December 2021). “The DREAM complex represses growth in response to DNA damage in Arabidopsis”. Life Science Alliance. 4 (12): e202101141. doi:10.26508/lsa.202101141. PMC 8500230. PMID 34583930.
- Sadasivam, S.; Duan, S.; DeCaprio, J. A. (5 March 2012). “The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression”. Genes & Development. 26 (5): 474–489. doi:10.1101/gad.181933.111. PMC 3305985. PMID 22391450.
- Litovchick, L; Florens, LA; Swanson, SK; Washburn, MP; DeCaprio, JA (2011-04-15). “DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly”. Genes & Development. 25 (8): 801–13. doi:10.1101/gad.2034211. PMC 3078706. PMID 21498570.
- Georlette, D; Ahn, S; MacAlpine, DM; Cheung, E; Lewis, PW; Beall, EL; Bell, SP; Speed, T; Manak, JR; Botchan, MR (2007-11-15). “Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells”. Genes & Development. 21 (22): 2880–96. doi:10.1101/gad.1600107. PMC 2049191. PMID 17978103.
- Rovani, Margritte K.; Brachmann, Carrie Baker; Ramsay, Gary; Katzen, Alisa L. (December 2012). “The dREAM/Myb–MuvB complex and Grim are key regulators of the programmed death of neural precursor cells at the Drosophila posterior wing margin”. Developmental Biology. 372 (1): 88–102. doi:10.1016/j.ydbio.2012.08.022. PMC 3621911. PMID 22960039.
- Müller, GA; Engeland, K (February 2010). “The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription”. The FEBS Journal. 277 (4): 877–93. doi:10.1111/j.1742-4658.2009.07508.x. PMID 20015071. S2CID 8955433.
- Forristal, C; Henley, SA; MacDonald, JI; Bush, JR; Ort, C; Passos, DT; Talluri, S; Ishak, CA; Thwaites, MJ; Norley, CJ; Litovchick, L; DeCaprio, JA; DiMattia, G; Holdsworth, DW; Beier, F; Dick, FA (June 2014). “Loss of the mammalian DREAM complex deregulates chondrocyte proliferation”. Molecular and Cellular Biology. 34 (12): 2221–34. doi:10.1128/MCB.01523-13. PMC 4054284. PMID 24710275.
- Guiley, KZ; Liban, TJ; Felthousen, JG; Ramanan, P; Litovchick, L; Rubin, SM (2015-05-01). “Structural mechanisms of DREAM complex assembly and regulation”. Genes & Development. 29 (9): 961–74. doi:10.1101/gad.257568.114. PMC 4421984. PMID 25917549.
- Sandoval, R; Pilkinton, M; Colamonici, OR (2009-10-15). “Deletion of the p107/p130-binding domain of Mip130/LIN-9 bypasses the requirement for CDK4 activity for the dissociation of Mip130/LIN-9 from p107/p130-E2F4 complex”. Experimental Cell Research. 315 (17): 2914–20. doi:10.1016/j.yexcr.2009.07.014. PMC 2757496. PMID 19619530.
- Stiegler, P; De Luca, A; Bagella, L; Giordano, A (1998-11-15). “The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter”. Cancer Research. 58 (22): 5049–52. PMID 9823308.
- Bai, L; Morozov, AV (November 2010). “Gene regulation by nucleosome positioning”. Trends in Genetics. 26 (11): 476–83. doi:10.1016/j.tig.2010.08.003. PMID 20832136.
- Latorre, I; Chesney, MA; Garrigues, JM; Stempor, P; Appert, A; Francesconi, M; Strome, S; Ahringer, J (2015-03-01). “The DREAM complex promotes gene body H2A.Z for target repression”. Genes & Development. 29 (5): 495–500. doi:10.1101/gad.255810.114. PMC 4358402. PMID 25737279.
- Tedesco, D; Lukas, J; Reed, SI (2002-11-15). “The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2)”. Genes & Development. 16 (22): 2946–57. doi:10.1101/gad.1011202. PMC 187481. PMID 12435635.
- Bhattacharya, S; Garriga, J; Calbó, J; Yong, T; Haines, DS; Graña, X (2003-04-24). “SKP2 associates with p130 and accelerates p130 ubiquitylation and degradation in human cells”. Oncogene. 22 (16): 2443–51. doi:10.1038/sj.onc.1206339. PMID 12717421. S2CID 26125392.
- Gaubatz, S; Lees, JA; Lindeman, GJ; Livingston, DM (February 2001). “E2F4 is exported from the nucleus in a CRM1-dependent manner”. Molecular and Cellular Biology. 21 (4): 1384–92. doi:10.1128/MCB.21.4.1384-1392.2001. PMC 99590. PMID 11158323.
- Takahashi, Y; Rayman, JB; Dynlacht, BD (2000-04-01). “Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression”. Genes & Development. 14 (7): 804–16. doi:10.1101/gad.14.7.804. PMC 316494. PMID 10766737.
- Lam, EW; Robinson, C; Watson, RJ (September 1992). “Characterization and cell cycle-regulated expression of mouse B-myb”. Oncogene. 7 (9): 1885–90. PMID 1501895.
- Pilkinton, M; Sandoval, R; Song, J; Ness, SA; Colamonici, OR (2007-01-05). “Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1”. The Journal of Biological Chemistry. 282 (1): 168–75. doi:10.1074/jbc.M609924200. PMID 17098733. S2CID 21963932.
- Kolupaeva, V; Janssens, V (January 2013). “PP1 and PP2A phosphatases–cooperating partners in modulating retinoblastoma protein activation”. The FEBS Journal. 280 (2): 627–43. doi:10.1111/j.1742-4658.2012.08511.x. PMID 22299668. S2CID 46705471.
- Kurimchak, A; Graña, X (2015). “PP2A: more than a reset switch to activate pRB proteins during the cell cycle and in response to signaling cues”. Cell Cycle. 14 (1): 18–30. doi:10.4161/15384101.2014.985069. PMC 4612414. PMID 25483052.
- Naetar, N; Soundarapandian, V; Litovchick, L; Goguen, KL; Sablina, AA; Bowman-Colin, C; Sicinski, P; Hahn, WC; DeCaprio, JA; Livingston, DM (2014-06-19). “PP2A-mediated regulation of Ras signaling in G2 is essential for stable quiescence and normal G1 length”. Molecular Cell. 54 (6): 932–45. doi:10.1016/j.molcel.2014.04.023. PMC 4118046. PMID 24857551.
- Chen, JY; Lin, JR; Tsai, FC; Meyer, T (2013-10-10). “Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle”. Molecular Cell. 52 (1): 87–100. doi:10.1016/j.molcel.2013.09.009. PMC 4039290. PMID 24119401.
- Wolter, P; Schmitt, K; Fackler, M; Kremling, H; Probst, L; Hauser, S; Gruss, OJ; Gaubatz, S (15 May 2012). “GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability”. Journal of Cell Science. 125 (Pt 10): 2393–406. doi:10.1242/jcs.097253. PMID 22344256.
- DeCaprio, James A.; Duensing, Anette (July 2014). “The DREAM complex in antitumor activity of imatinib mesylate in gastrointestinal stromal tumors”. Current Opinion in Oncology. 26 (4): 415–421. doi:10.1097/CCO.0000000000000090. PMC 4236229. PMID 24840522.
- Boichuk, S.; Parry, J. A.; Makielski, K. R.; Litovchick, L.; Baron, J. L.; Zewe, J. P.; Wozniak, A.; Mehalek, K. R.; Korzeniewski, N.; Seneviratne, D. S.; Schoffski, P.; Debiec-Rychter, M.; DeCaprio, J. A.; Duensing, A. (20 June 2013). “The DREAM Complex Mediates GIST Cell Quiescence and Is a Novel Therapeutic Target to Enhance Imatinib-Induced Apoptosis”. Cancer Research. 73 (16): 5120–5129. doi:10.1158/0008-5472.CAN-13-0579. PMID 23786773.
Further reading
- Rashid, NN; Yusof, R; Watson, RJ (November 2014). “A B-myb–DREAM complex is not critical to regulate the G2/M genes in HPV-transformed cell lines”. Anticancer Research. 34 (11): 6557–63. PMID 25368258.
from see also entries
- Faha B, Ewen ME, Tsai LH, Livingston DM, Harlow E (Jan 1992). “Interaction between human cyclin A and adenovirus E1A-associated p107 protein”. Science. 255 (5040): 87–90. Bibcode:1992Sci…255…87F. doi:10.1126/science.1532458. PMID 1532458.
- Ewen ME, Xing YG, Lawrence JB, Livingston DM (Sep 1991). “Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein”. Cell. 66 (6): 1155–64. doi:10.1016/0092-8674(91)90038-Z. PMID 1833063. S2CID 27478008.
- Datta PK, Raychaudhuri P, Bagchi S (Oct 1995). “Association of p107 with Sp1: genetically separable regions of p107 are involved in regulation of E2F- and Sp1-dependent transcription”. Molecular and Cellular Biology. 15 (10): 5444–52. doi:10.1128/mcb.15.10.5444. PMC 230794. PMID 7565695.
- Zhu L, Zhu L, Xie E, Chang LS (Jul 1995). “Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins”. Molecular and Cellular Biology. 15 (7): 3552–62. doi:10.1128/mcb.15.7.3552. PMC 230592. PMID 7791762.
- Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg RA (Mar 1995). “E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle”. Proceedings of the National Academy of Sciences of the United States of America. 92 (6): 2403–7. Bibcode:1995PNAS…92.2403S. doi:10.1073/pnas.92.6.2403. PMC 42492. PMID 7892279.
- Kim YW, Otterson GA, Kratzke RA, Coxon AB, Kaye FJ (Nov 1994). “Differential specificity for binding of retinoblastoma binding protein 2 to RB, p107, and TATA-binding protein”. Molecular and Cellular Biology. 14 (11): 7256–64. doi:10.1128/mcb.14.11.7256. PMC 359260. PMID 7935440.
- Ginsberg D, Vairo G, Chittenden T, Xiao ZX, Xu G, Wydner KL, DeCaprio JA, Lawrence JB, Livingston DM (Nov 1994). “E2F-4, a new member of the E2F transcription factor family, interacts with p107”. Genes & Development. 8 (22): 2665–79. doi:10.1101/gad.8.22.2665. PMID 7958924.
- Beijersbergen RL, Kerkhoven RM, Zhu L, Carlée L, Voorhoeve PM, Bernards R (Nov 1994). “E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo”. Genes & Development. 8 (22): 2680–90. doi:10.1101/gad.8.22.2680. PMID 7958925.
- Beijersbergen RL, Hijmans EM, Zhu L, Bernards R (Sep 1994). “Interaction of c-Myc with the pRb-related protein p107 results in inhibition of c-Myc-mediated transactivation”. The EMBO Journal. 13 (17): 4080–6. doi:10.1002/j.1460-2075.1994.tb06725.x. PMC 395329. PMID 8076603.
- Dyson N, Dembski M, Fattaey A, Ngwu C, Ewen M, Helin K (Dec 1993). “Analysis of p107-associated proteins: p107 associates with a form of E2F that differs from pRB-associated E2F-1”. Journal of Virology. 67 (12): 7641–7. doi:10.1128/JVI.67.12.7641-7647.1993. PMC 238233. PMID 8230483.
- Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E (Jul 1993). “Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein”. Genes & Development. 7 (7A): 1111–25. doi:10.1101/gad.7.7a.1111. PMID 8319904.
- Ikeda MA, Jakoi L, Nevins JR (Apr 1996). “A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation”. Proceedings of the National Academy of Sciences of the United States of America. 93 (8): 3215–20. Bibcode:1996PNAS…93.3215I. doi:10.1073/pnas.93.8.3215. PMC 39585. PMID 8622916.
- Xiao ZX, Ginsberg D, Ewen M, Livingston DM (May 1996). “Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases”. Proceedings of the National Academy of Sciences of the United States of America. 93 (10): 4633–7. Bibcode:1996PNAS…93.4633X. doi:10.1073/pnas.93.10.4633. PMC 39330. PMID 8643455.
- Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD (Sep 1996). “Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions”. Proceedings of the National Academy of Sciences of the United States of America. 93 (19): 10315–20. Bibcode:1996PNAS…9310315V. doi:10.1073/pnas.93.19.10315. PMC 38381. PMID 8816797.
- Shao Z, Siegert JL, Ruppert S, Robbins PD (Jul 1997). “Rb interacts with TAF(II)250/TFIID through multiple domains”. Oncogene. 15 (4): 385–92. doi:10.1038/sj.onc.1201204. PMID 9242374.
- Verona R, Moberg K, Estes S, Starz M, Vernon JP, Lees JA (Dec 1997). “E2F activity is regulated by cell cycle-dependent changes in subcellular localization”. Molecular and Cellular Biology. 17 (12): 7268–82. doi:10.1128/mcb.17.12.7268. PMC 232584. PMID 9372959.
- Trimarchi JM, Fairchild B, Verona R, Moberg K, Andon N, Lees JA (Mar 1998). “E2F-6, a member of the E2F family that can behave as a transcriptional repressor”. Proceedings of the National Academy of Sciences of the United States of America. 95 (6): 2850–5. Bibcode:1998PNAS…95.2850T. doi:10.1073/pnas.95.6.2850. PMC 19658. PMID 9501179.
- Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM (May 1998). “Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7”. Molecular and Cellular Biology. 18 (5): 2748–57. doi:10.1128/mcb.18.5.2748. PMC 110654. PMID 9566894.
- Woitach JT, Zhang M, Niu CH, Thorgeirsson SS (Aug 1998). “A retinoblastoma-binding protein that affects cell-cycle control and confers transforming ability”. Nature Genetics. 19 (4): 371–4. doi:10.1038/1258. PMID 9697699. S2CID 11374970.
- Veal E, Eisenstein M, Tseng ZH, Gill G (Sep 1998). “A cellular repressor of E1A-stimulated genes that inhibits activation by E2F”. Molecular and Cellular Biology. 18 (9): 5032–41. doi:10.1128/mcb.18.9.5032. PMC 109088. PMID 9710587.
- Soprano KJ, Purev E, Vuocolo S, Soprano DR (Aug 2006). “Rb2/p130 and protein phosphatase 2A: key mediators of ovarian carcinoma cell growth suppression by all-trans retinoic acid”. Oncogene. 25 (38): 5315–25. doi:10.1038/sj.onc.1209679. PMID 16936753. S2CID 19893445.
- De Falco G, Giordano A (Aug 2006). “pRb2/p130: a new candidate for retinoblastoma tumor formation”. Oncogene. 25 (38): 5333–40. doi:10.1038/sj.onc.1209614. PMID 16936755. S2CID 40396708.
- Hijmans EM, Voorhoeve PM, Beijersbergen RL, van ‘t Veer LJ, Bernards R (Jun 1995). “E2F-5, a new E2F family member that interacts with p130 in vivo”. Molecular and Cellular Biology. 15 (6): 3082–9. doi:10.1128/mcb.15.6.3082. PMC 230539. PMID 7760804.
- Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg RA (Mar 1995). “E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle”. Proceedings of the National Academy of Sciences of the United States of America. 92 (6): 2403–7. Bibcode:1995PNAS…92.2403S. doi:10.1073/pnas.92.6.2403. PMC 42492. PMID 7892279.
- Yeung RS, Bell DW, Testa JR, Mayol X, Baldi A, Graña X, Klinga-Levan K, Knudson AG, Giordano A (Dec 1993). “The retinoblastoma-related gene, RB2, maps to human chromosome 16q12 and rat chromosome 19”. Oncogene. 8 (12): 3465–8. PMID 8247552.
- Li Y, Graham C, Lacy S, Duncan AM, Whyte P (Dec 1993). “The adenovirus E1A-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E”. Genes & Development. 7 (12A): 2366–77. doi:10.1101/gad.7.12a.2366. PMID 8253383.
- Hannon GJ, Demetrick D, Beach D (Dec 1993). “Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins”. Genes & Development. 7 (12A): 2378–91. doi:10.1101/gad.7.12a.2378. PMID 8253384.
- Lavender P, Vandel L, Bannister AJ, Kouzarides T (Jun 1997). “The HMG-box transcription factor HBP1 is targeted by the pocket proteins and E1A”. Oncogene. 14 (22): 2721–8. doi:10.1038/sj.onc.1201243. PMID 9178770.
- Lacy S, Whyte P (May 1997). “Identification of a p130 domain mediating interactions with cyclin A/cdk 2 and cyclin E/cdk 2 complexes”. Oncogene. 14 (20): 2395–406. doi:10.1038/sj.onc.1201085. PMID 9188854. S2CID 26359262.
- Shao Z, Siegert JL, Ruppert S, Robbins PD (Jul 1997). “Rb interacts with TAF(II)250/TFIID through multiple domains”. Oncogene. 15 (4): 385–92. doi:10.1038/sj.onc.1201204. PMID 9242374.
- Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM (May 1998). “Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7”. Molecular and Cellular Biology. 18 (5): 2748–57. doi:10.1128/mcb.18.5.2748. PMC 110654. PMID 9566894.
- Woitach JT, Zhang M, Niu CH, Thorgeirsson SS (Aug 1998). “A retinoblastoma-binding protein that affects cell-cycle control and confers transforming ability”. Nature Genetics. 19 (4): 371–4. doi:10.1038/1258. PMID 9697699. S2CID 11374970.
- Veal E, Eisenstein M, Tseng ZH, Gill G (Sep 1998). “A cellular repressor of E1A-stimulated genes that inhibits activation by E2F”. Molecular and Cellular Biology. 18 (9): 5032–41. doi:10.1128/mcb.18.9.5032. PMC 109088. PMID 9710587.
- Fusco C, Reymond A, Zervos AS (Aug 1998). “Molecular cloning and characterization of a novel retinoblastoma-binding protein”. Genomics. 51 (3): 351–8. doi:10.1006/geno.1998.5368. PMID 9721205.
- Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D (Sep 1998). “The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase”. Proceedings of the National Academy of Sciences of the United States of America. 95 (18): 10493–8. Bibcode:1998PNAS…9510493F. doi:10.1073/pnas.95.18.10493. PMC 27922. PMID 9724731.
- Paramio JM, Laín S, Segrelles C, Lane EB, Jorcano JL (Aug 1998). “Differential expression and functionally co-operative roles for the retinoblastoma family of proteins in epidermal differentiation”. Oncogene. 17 (8): 949–57. doi:10.1038/sj.onc.1202031. PMID 9747874. S2CID 21804961.
- Wang S, Ghosh RN, Chellappan SP (Dec 1998). “Raf-1 physically interacts with Rb and regulates its function: a link between mitogenic signaling and cell cycle regulation”. Molecular and Cellular Biology. 18 (12): 7487–98. doi:10.1128/mcb.18.12.7487. PMC 109329. PMID 9819434.
- Yang R, Müller C, Huynh V, Fung YK, Yee AS, Koeffler HP (Mar 1999). “Functions of cyclin A1 in the cell cycle and its interactions with transcription factor E2F-1 and the Rb family of proteins”. Molecular and Cellular Biology. 19 (3): 2400–7. doi:10.1128/mcb.19.3.2400. PMC 84032. PMID 10022926.


Leave a Reply