Steroidogenic factor 1 (SF-1) protein and a few related things
The steroidogenic factor 1 (SF-1) protein is a transcription factor involved in sex determination by controlling the activity of genes related to the reproductive glands or gonads and adrenal glands. This protein is encoded by the NR5A1 gene, a member of the nuclear receptor subfamily, located on the long arm of chromosome 9 at position 33.3. It was originally identified as a regulator of genes encoding cytochrome P450 steroid hydroxylases, however, further roles in endocrine function have since been discovered.
- Reference, Genetics Home. “NR5A1 gene”. Genetics Home Reference. Retrieved 2017-11-30.
- Parker KL, Schimmer BP (June 1997). “Steroidogenic factor 1: a key determinant of endocrine development and function”. Endocrine Reviews. 18 (3): 361–77. doi:10.1210/edrv.18.3.0301. PMID 9183568.
Structure
The NR5A1 gene encodes a 461-amino acid protein that shares several conserved domains consistent with members of the nuclear receptor subfamily. The N-terminal domain includes two zinc fingers and is responsible for DNA binding via specific recognition of target sequences. Variations of AGGTCA DNA motifs allows SF-1 to interact with the major groove of the DNA helix and monomerically bind. Following binding, trans-activation of target genes depends on recruitment of co-activators such as SRC-1, GRIP1, PNRC, or GCN5. Other critical domains of SF-1 include a proline-rich hinge region, ligand-binding domain, and a C-terminal activation domain for transcriptional interactions. A 30-amino acid extension of the DNA-binding domain known as the A-box stabilizes monomeric binding by acting as a DNA anchor. The hinge region can undergo post-transcriptional and translational modifications such as phosphorylation by cAMP-dependent kinase, that further enhance stability and transcriptional activity.
- Parker KL, Schimmer BP (June 1997). “Steroidogenic factor 1: a key determinant of endocrine development and function”. Endocrine Reviews. 18 (3): 361–77. doi:10.1210/edrv.18.3.0301. PMID 9183568.
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM (December 1995). “The nuclear receptor superfamily: the second decade”. Cell. 83 (6): 835–9. doi:10.1016/0092-8674(95)90199-x. PMC 6159888. PMID 8521507.
- Honda S, Morohashi K, Nomura M, Takeya H, Kitajima M, Omura T (April 1993). “Ad4BP regulating steroidogenic P-450 gene is a member of steroid hormone receptor superfamily”. The Journal of Biological Chemistry. 268 (10): 7494–502. doi:10.1016/S0021-9258(18)53202-6. PMID 8463279.
SF-1 is considered an orphan receptor as high-affinity naturally occurring ligands have yet to be identified.
Homology
Analysis of mouse SF-1 cDNA revealed sequence similarities with Drosophila fushi tarazu factor I (FTZ-F1) which regulates the fushi tarazu homeobox gene. Several other FTZ-F1 homologs have been identified that implicate high level of sequence conservation among vertebrates and invertebrates. For example, SF-1 cDNA shares an identical 1017 base-pair sequence with embryonal long terminal repeat-binding protein (ELP) cDNA isolated from embryonal carcinoma cells, differing only in their terminal ends.
- Ikeda Y, Lala DS, Luo X, Kim E, Moisan MP, Parker KL (July 1993). “Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression”. Molecular Endocrinology. 7 (7): 852–60. doi:10.1210/mend.7.7.8413309. PMID 8413309.
Expression
Adult steroidogenic tissue
SF-1 expression is localized to adult steroidogenic tissues correlating with known expression profiles of steroid hydroxylases. Using in situ hybridization with SF-1 cRNA specific probe detected gene transcripts in adrenocortical cells, Leydig cells, and ovarian theca and granulosa cells. SF-1 specific antibody studies confirmed expression profile of SF-1 in rats and humans corresponding to sites of transcript detection.
- Ikeda Y, Lala DS, Luo X, Kim E, Moisan MP, Parker KL (July 1993). “Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression”. Molecular Endocrinology. 7 (7): 852–60. doi:10.1210/mend.7.7.8413309. PMID 8413309.
- Morohashi K, Iida H, Nomura M, Hatano O, Honda S, Tsukiyama T, Niwa O, Hara T, Takakusu A, Shibata Y (May 1994). “Functional difference between Ad4BP and ELP, and their distributions in steroidogenic tissues”. Molecular Endocrinology. 8 (5): 643–53. doi:10.1210/mend.8.5.8058072. PMID 8058072.
- Takayama K, Sasano H, Fukaya T, Morohashi K, Suzuki T, Tamura M, Costa MJ, Yajima A (September 1995). “Immunohistochemical localization of Ad4-binding protein with correlation to steroidogenic enzyme expression in cycling human ovaries and sex cord stromal tumors”. The Journal of Clinical Endocrinology and Metabolism. 80 (9): 2815–21. doi:10.1210/jcem.80.9.7673429. PMID 7673429.
Embryonic steroidogenic tissue
Genetic sex in mammals is determined by the presence or absence of the Y chromosome at fertilization. Sexually dimorphic development of embryonic gonads into testes or ovaries is activated by the SRY gene product. Sexual differentiation is then directed by hormones produced by embryonic testes, the presence of ovaries, or complete absence of gonads. SF-1 transcripts initially localize to the urogenital ridge before SF-1 expressing cells resolve into distinct adrenocortical and gonadal precursors that ultimately give rise to adrenal cortex and gonads.
- “Male Development of Chromosomally Female Mice Transgenic for Sry gene” (1991), by Peter Koopman, et al. | The Embryo Project Encyclopedia”. embryo.asu.edu. Retrieved 2017-11-30.
SF-1 transcripts precede the onset of SRY expression in the fetal testes hinting at gonadal developmental role. SRY influences the differentiation of the fetal testes into distinct compartments: testicular cords and interstitial region containing Leydig cells. Increase in SF-1 protein and detection in the steroidogenic Leydig cells and testicular cords coincides with development.
- “Male Development of Chromosomally Female Mice Transgenic for Sry gene” (1991), by Peter Koopman, et al. | The Embryo Project Encyclopedia”. embryo.asu.edu. Retrieved 2017-11-30.
However, in the ovaries, gonadal sexual differentiation is facilitated by reductions in SF-1 transcript and protein. SF-1 levels is strongly expressed at the onset of follicular development in theca and granulosa cells which precedes expression of the aromatase enzyme responsible for estrogen biosynthesis.
Other sites
Embryonic mouse SF-1 transcripts have been discovered to localize within regions of the developing diencephalon and subsequently in the ventromedial hypothalamic nucleus (VMH) suggesting roles beyond steroidogenic maintenance.
- Ikeda Y, Lala DS, Luo X, Kim E, Moisan MP, Parker KL (July 1993). “Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression”. Molecular Endocrinology. 7 (7): 852–60. doi:10.1210/mend.7.7.8413309. PMID 8413309.
The ventromedial nucleus of the hypothalamus (VMN, VMH or ventromedial hypothalamus) is a nucleus of the hypothalamus. In 2007, Kurrasch et al. found that the ventromedial hypothalamus is a distinct morphological nucleus involved in terminating hunger, fear, thermoregulation, and sexual activity.
- Kurrasch DM, Cheung CC, Lee FY, Tran PV, Hata K, Ingraham HA (December 2007). “The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning”. The Journal of Neuroscience. 27 (50): 13624–34. doi:10.1523/JNEUROSCI.2858-07.2007. PMC 6673626. PMID 18077674.
This nuclear region is involved in the recognition of the feeling of fullness.
It has four subdivisions:
- Anterior (VMHa)
- Dorsomedial (VMHdm)
- Ventrolateral (VMHvl)
- Central (VMHc)
These subdivisions differ anatomically, neurochemically, and behaviorally.
The ventromedial nucleus (VMN) is most commonly associated with satiety. Early studies showed that VMN lesions caused over-eating and obesity in rats. However, the interpretation of these experiments was summarily discredited when Gold’s research demonstrated that precision lesioning of the VMN did not result in hyperphagia. Nevertheless, numerous studies have shown that the immediacy of hyperphagia and obesity syndrome are a consequence of VMN lesions or procaine injections, and point to the VMN’s role in satiety. A major review of the subject in 2006 concluded that, “anatomical studies done both before and after Gold’s study did not replicate his results with lesions, and in nearly every published direct comparison of VMH lesions vs. PVN or VNAB lesions, the group with VMH lesions ate substantially more food and gained twice as much weight.” This strongly substantiates the classification of VMN as the primary satiety center in the hypothalamus.
- Gold RM (November 1973). “Hypothalamic obesity: the myth of the ventromedial nucleus”. Science. 182 (4111): 488–90. Bibcode:1973Sci…182..488G. doi:10.1126/science.182.4111.488. PMID 4795550. S2CID 3011420.
- Balagura S, Devenport LD (June 1970). “Feeding patterns of normal and ventromedial hypothalamic lesioned male and female rats”. Journal of Comparative and Physiological Psychology. 71 (3): 357–64. doi:10.1037/h0029118. PMID 5480868.
- Becker EE, Kissileff HR (February 1974). “Inhibitory controls of feeding by the ventromedial hypothalamus”. The American Journal of Physiology. 226 (2): 383–96. doi:10.1152/ajplegacy.1974.226.2.383. PMID 4811195.
- Berthoud HR, Jeanrenaud B (September 1979). “Changes of insulinemia, glycemia and feeding behavior induced by VMH-procainization in the rat”. Brain Research. 174 (1): 184–7. doi:10.1016/0006-8993(79)90816-3. PMID 487120. S2CID 39015121.
- Brooks CM, Lockwood RA, Wiggins ML (December 1946). “A study of the effect of hypothalamic lesions on the eating habits of the albino rat”. The American Journal of Physiology. 147 (4): 735–41. doi:10.1152/ajplegacy.1946.147.4.735. PMID 20277066.
- Epstein AN (December 1960). “Reciprocal changes in feeding behavior produced by intrahypothalamic chemical injections”. The American Journal of Physiology. 199 (6): 969–74. doi:10.1152/ajplegacy.1960.199.6.969. PMID 13697000.
- Larkin RP (November 1975). “Effect of ventromedial hypothalamic procaine injections on feeding, lever pressing, and other behavior in rats”. Journal of Comparative and Physiological Psychology. 89 (9): 1100–8. doi:10.1037/h0077192. PMID 1202103.
- Maes H (June 1980). “Time course of feeding induced by pentobarbital-injections into the rat’s VMH”. Physiology & Behavior. 24 (6): 1107–14. doi:10.1016/0031-9384(80)90055-4. PMID 7413790. S2CID 43051882.
- King BM (February 2006). “The rise, fall, and resurrection of the ventromedial hypothalamus in the regulation of feeding behavior and body weight”. Physiology & Behavior. 87 (2): 221–44. doi:10.1016/j.physbeh.2005.10.007. PMID 16412483. S2CID 40880350.
It has also been found that lesions to the VMH in rats caused increased plasma insulin levels. Rats with a VMH lesion compared to normal rats overproduce a circulating satiety factor, to which the control rats can respond and rats with a VMH lesion cannot respond. A lesion to the VMH makes rats overproduce leptin, which they cannot respond to causing them to over eat, leading to obesity.
- Satoh N, Ogawa Y, Katsuura G, Tsuji T, Masuzaki H, Hiraoka J, Okazaki T, Tamaki M, Hayase M, Yoshimasa Y, Nishi S, Hosoda K, Nakao K (March 1997). “Pathophysiological significance of the obese gene product, leptin, in ventromedial hypothalamus (VMH)-lesioned rats: evidence for loss of its satiety effect in VMH-lesioned rats”. Endocrinology. 138 (3): 947–54. doi:10.1210/endo.138.3.4989. PMID 9048594.
Researchers looked at a series of twenty-one animals of various degrees of adiposity, with respect to growth appearance, fat distribution, general physical condition, and the correlation between the level of adiposity attained and the correlation of the hypothalamic lesion. Lesions in the hypothalamic area, particularly the region of the ventromedial hypothalamus interrupts a large number of the descending fibers from the hypothalamic cell groups that were found to contribute to obesity in rats.
- Hetherington AW, Ranson SW (June 1942). “The relation of various hypothalamic lesions to adiposity in the rat” (PDF). Journal of Comparative Neurology. 76 (3): 475–99. doi:10.1002/cne.900760308. S2CID 85715802.
Another study found that there seems to be a higher concentration of cannabinoid receptor mRNA within the VMH in comparison to other nuclei within the hypothalamus. The cannabinoid ingestion has been linked to rewarding processes, and also with the release of dopamine in the brain.
- Jamshidi N, Taylor DA (November 2001). “Anandamide administration into the ventromedial hypothalamus stimulates appetite in rats”. British Journal of Pharmacology. 134 (6): 1151–4. doi:10.1038/sj.bjp.0704379. PMC 1573067. PMID 11704633.
VMH is also important in mammal play behaviour. Lesions to VMH along with the hippocampus, amygdala, the cerebellum, and the lateral hypothalamus are all linked to reduced play.
- Panksepp J, Siviy S, Normansell L (1984). “The psychobiology of play: theoretical and methodological perspectives”. Neuroscience and Biobehavioral Reviews. 8 (4): 465–92. doi:10.1016/0149-7634(84)90005-8. PMID 6392950. S2CID 26810046.
The VMHdm has a role in the male vocalizations and scent marking behaviors.
- Flanagan-Cato LM, Lee BJ, Calizo LH (June 2006). “Co-localization of midbrain projections, progestin receptors, and mating-induced fos in the hypothalamic ventromedial nucleus of the female rat”. Hormones and Behavior. 50 (1): 52–60. doi:10.1016/j.yhbeh.2006.01.012. PMID 16546183. S2CID 36201218.
- Harding SM, McGinnis MY (October 2005). “Microlesions of the ventromedial nucleus of the hypothalamus: effects on sociosexual behaviors in male rats”. Behavioral Neuroscience. 119 (5): 1227–34. doi:10.1037/0735-7044.119.5.1227. PMID 16300430.
The VMHvl contains many distinct neuronal populations that contribute to varying, often distinct, functions. Notably, this region plays a role in sexual behaviors in females (lordosis), thus stimulating their sexual arousal. The VMHvl has also been found to play a role in estrogen-mediated movement and energy expenditure/thermogenesis.
- Kammel LG, Correa SM (January 2020). “Selective sexual differentiation of neurone populations may contribute to sex-specific outputs of the ventromedial nucleus of the hypothalamus”. Journal of Neuroendocrinology. 32 (1): e12801. doi:10.1111/jne.12801. PMC 6982598. PMID 31605642.
- Kow LM, Pfaff DW (May 1998). “Mapping of neural and signal transduction pathways for lordosis in the search for estrogen actions on the central nervous system”. Behavioural Brain Research. 92 (2): 169–80. doi:10.1016/S0166-4328(97)00189-7. PMID 9638959. S2CID 28276218.
- Christensen LW, Nance DM, Gorski RA (1977). “Effects of hypothalamic and preoptic lesions on reproductive behavior in male rats”. Brain Research Bulletin. 2 (2): 137–41. doi:10.1016/0361-9230(77)90010-7. PMID 880486. S2CID 4700161.
- Pfaff DW, Sakuma Y (March 1979). “Facilitation of the lordosis reflex of female rats from the ventromedial nucleus of the hypothalamus”. The Journal of Physiology. 288: 189–202. doi:10.1113/jphysiol.1979.sp012690. PMC 1281421. PMID 469715.
- Matsumoto T, Yamanouchi K (September 2000). “Acceleration of mounting behaviors in female rats by ibotenic acid lesions in the ventromedial hypothalamic nucleus”. Neuroscience Letters. 291 (3): 143–6. doi:10.1016/S0304-3940(00)01388-4. PMID 10984627. S2CID 10334038.
- Correa SM, Newstrom DW, Warne JP, Flandin P, Cheung CC, Lin-Moore AT, Pierce AA, Xu AW, Rubenstein JL, Ingraham HA (January 2015). “An Estrogen-Responsive Module in the Ventromedial Hypothalamus Selectively Drives Sex-Specific Activity in Females”. Cell Reports. 10 (1): 62–74. doi:10.1016/j.celrep.2014.12.011. PMC 4324838. PMID 25543145.
- van Veen JE, Kammel LG, Bunda PC, Shum M, Reid MS, Massa MG, Arneson D, Park JW, Zhang Z, Joseph AM, Hrncir H, Liesa M, Arnold AP, Yang X, Correa SM (April 2020). “Hypothalamic estrogen receptor alpha establishes a sexually dimorphic regulatory node of energy expenditure”. Nature Metabolism. 2 (4): 351–63. doi:10.1038/s42255-020-0189-6. PMC 7202561. PMID 32377634.
Bilateral FOS expression in the VMH after repeated seizures is associated with alteration in the severity of flurothyl induced seizures in C57BL/6J mice that are not present in DBA/2J mice. Moreover, bilateral lesions of the VMH are able to block the propagation of seizure discharge to enter the brainstem seizure system.
- Kadiyala SB, Papandrea D, Tuz K, Anderson TM, Jayakumar S, Herron BJ, Ferland RJ (January 2015). “Spatiotemporal differences in the c-fos pathway between C57BL/6J and DBA/2J mice following flurothyl-induced seizures: A dissociation of hippocampal Fos from seizure activity”. Epilepsy Research. 109: 183–96. doi:10.1016/j.eplepsyres.2014.11.009. PMC 4272448. PMID 25524858.
- Kadiyala SB, Ferland RJ (March 2017). “Dissociation of spontaneous seizures and brainstem seizure thresholds in mice exposed to eight flurothyl-induced generalized seizures”. Epilepsia Open. 2 (1): 48–58. doi:10.1002/epi4.12031. PMC 5560332. PMID 28825051.
- Ferland RJ, Applegate CD (November 1998). “The role of the ventromedial nucleus of the hypothalamus in epileptogenesis”. NeuroReport. 9 (16): 3623–9. doi:10.1097/00001756-199811160-00013. PMID 9858370. S2CID 29713035.
Surgery
In West Germany, at least 70 men had their VMN operated on between 1962 and 1979. Most of these individuals had been involuntarily institutionalized or imprisoned for deviant sexual behavior, such as homosexuality, perceived hypersexuality among heterosexual men, and pedophilia. This surgery was not commonly performed elsewhere.
- Rieber, Inge; Sigusch, Volkmar (1979). “Psychosurgery on sex offenders and sexual ?deviants? in West Germany”. Archives of Sexual Behavior. 8 (6): 523–527. doi:10.1007/BF01541419. PMID 391177. S2CID 41463669.
RT-PCR approaches have detected transcripts of mice FTZ-F1 gene in the placenta and spleen; and SF-1 transcripts in the human placenta.
- Ninomiya Y, Okada M, Kotomura N, Suzuki K, Tsukiyama T, Niwa O (1995). “Genomic organization and isoforms of the mouse ELP gene”. Journal of Biochemistry. 118 (2): 380–9. doi:10.1093/oxfordjournals.jbchem.a124918. PMID 8543574.
Post-translational regulation
Transcription capacity of SF-1 can be influenced by post-translational modification. Specifically, phosphorylation of serine 203 is mediated by cyclin-dependent kinase 7. Mutations to CDK7 prevent interaction with the basal transcription factor, TFIIH, and formation of CDK-activating kinase complex. This inactivity has shown to repress phosphorylation of SF-1 and SF-1-dependent transcription.
- Lewis AE, Rusten M, Hoivik EA, Vikse EL, Hansson ML, Wallberg AE, Bakke M (January 2008). “Phosphorylation of steroidogenic factor 1 is mediated by cyclin-dependent kinase 7”. Molecular Endocrinology. 22 (1): 91–104. doi:10.1210/me.2006-0478. PMC 5419630. PMID 17901130.
Function
SF-1 is a critical regulator of reproduction, regulating the transcription of key genes involved in sexual development and reproduction, most notably StAR and P450SCC. It can form a transcriptional complex with TDF to up-regulate transcription of the Sox9 gene. Its targets include genes at every level of the hypothalamic-pituitary-gonadal axis, as well as many genes involved in gonadal and adrenal steroidogenesis.
- Jameson JL (December 2004). “Of mice and men: The tale of steroidogenic factor-1”. The Journal of Clinical Endocrinology and Metabolism. 89 (12): 5927–9. doi:10.1210/jc.2004-2047. PMID 15579738.
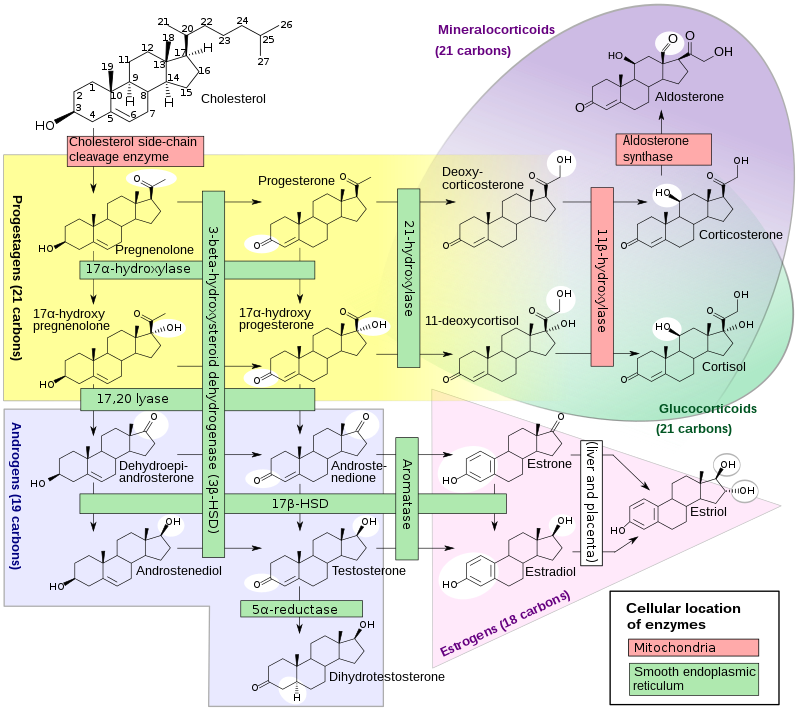
Cholesterol side-chain cleavage enzyme is commonly referred to as P450scc, where “scc” is an acronym for side-chaincleavage. P450scc is a mitochondrialenzyme that catalyzes conversion of cholesterol to pregnenolone. This is the first reaction in the process of steroidogenesis in all mammalian tissues that specialize in the production of various steroid hormones. P450scc is a member of the cytochrome P450 superfamily of enzymes (family 11, subfamily A, polypeptide 1) and is encoded by the CYP11A1 gene.
- Hanukoglu I (December 1992). “Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis”. The Journal of Steroid Biochemistry and Molecular Biology. 43 (8): 779–804. doi:10.1016/0960-0760(92)90307-5. PMID 22217824. S2CID 112729.
- “Entrez Gene: CYP11A1 cytochrome P450, family 11, subfamily A, polypeptide 1”.
The systematic name of this enzyme class is cholesterol, reduced-adrenal-ferredoxin:oxygen oxidoreductase (side-chain-cleaving). Other names include:
- C27-side-chain cleavage enzyme
- cholesterol 20-22-desmolase
- cholesterol C20-22 desmolase
- cholesterol desmolase
- cholesterol side-chain cleavage enzyme
- cholesterol side-chain-cleaving enzyme
- cytochrome P-450scc
- desmolase, steroid 20-22
- enzymes, cholesterol side-chain-cleaving
- steroid 20-22 desmolase
- steroid 20-22-lyase.
Tissue and intracellular localization
The highest level of the cholesterol side-chain cleavage system is found in the adrenal cortex and the corpus luteum. The system is also expressed at high levels in steroidogenic theca cells in the ovary, and Leydig cells in the testis. During pregnancy, the placenta also expresses significant levels of this enzyme system. P450scc is also present at much lower levels in several other tissue types, including the brain. In the adrenal cortex, the concentration of adrenodoxin is similar to that of P450scc, but adrenodoxin reductase is expressed at lower levels
- Hanukoglu I (December 1992). “Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis”. The Journal of Steroid Biochemistry and Molecular Biology. 43 (8): 779–804. doi:10.1016/0960-0760(92)90307-5. PMID 22217824. S2CID 112729.
- Strauss JF, Martinez F, Kiriakidou M (February 1996). “Placental steroid hormone synthesis: unique features and unanswered questions”. Biology of Reproduction. 54 (2): 303–311. doi:10.1095/biolreprod54.2.303. PMID 8788180.
- Stoffel-Wagner B (December 2001). “Neurosteroid metabolism in the human brain”. European Journal of Endocrinology. 145 (6): 669–679. doi:10.1530/eje.0.1450669. PMID 11720889.
- Hanukoglu I, Hanukoglu Z (May 1986). “Stoichiometry of mitochondrial cytochromes P-450, adrenodoxin and adrenodoxin reductase in adrenal cortex and corpus luteum. Implications for membrane organization and gene regulation”. European Journal of Biochemistry. 157 (1): 27–31. doi:10.1111/j.1432-1033.1986.tb09633.x. PMID 3011431.
Immunofluorescence studies using specific antibodies against P450scc system enzymes have demonstrated that proteins are located exclusively within the mitochondria. P450scc is associated with the inner mitochondrial membrane, facing the interior (matrix). Adrenodoxin and adrenodoxin reductase are soluble peripheral membrane proteins located inside the mitochondrial matrix that appear to associate with each other primarily through electrostatic interactions.
- Hanukoglu I, Suh BS, Himmelhoch S, Amsterdam A (October 1990). “Induction and mitochondrial localization of cytochrome P450scc system enzymes in normal and transformed ovarian granulosa cells”. The Journal of Cell Biology. 111 (4): 1373–1381. doi:10.1083/jcb.111.4.1373. PMC 2116250. PMID 2170421.
- Hanukoglu I, Feuchtwanger R, Hanukoglu A (November 1990). “Mechanism of corticotropin and cAMP induction of mitochondrial cytochrome P450 system enzymes in adrenal cortex cells”. The Journal of Biological Chemistry. 265 (33): 20602–20608. doi:10.1016/S0021-9258(17)30545-8. PMID 2173715.
- Topological studies of cytochromes P-450scc and P-45011 beta in bovine adrenocortical inner mitochondrial membranes. Effects of controlled tryptic digestion. J. Biol. Chem. 1979 254: 10443-8.
- Farkash Y, Timberg R, Orly J (April 1986). “Preparation of antiserum to rat cytochrome P-450 cholesterol side chain cleavage, and its use for ultrastructural localization of the immunoreactive enzyme by protein A-gold technique”. Endocrinology. 118 (4): 1353–1365. doi:10.1210/endo-118-4-1353. PMID 3948785.
- Hanukoglu I, Privalle CT, Jefcoate CR (May 1981). “Mechanisms of ionic activation of adrenal mitochondrial cytochromes P-450scc and P-45011 beta”. The Journal of Biological Chemistry. 256 (9): 4329–4335. doi:10.1016/S0021-9258(19)69437-8. PMID 6783659.
Mechanism of action
P450scc catalyzes the conversion of cholesterol to pregnenolone in three monooxygenase reactions. These involve 2 hydroxylations of the cholesterol side-chain, which generate, first, 22R-hydroxycholesterol and then 20alpha,22R-dihydroxycholesterol. The final step cleaves the bond between carbons 20 and 22, resulting in the production of pregnenolone and isocaproic aldehyde.
Each monooxygenase step requires 2 electrons (reducing equivalents). The initial source of the electrons is NADPH. The electrons are transferred from NADPH to P450scc via two electron transfer proteins: adrenodoxin reductase and adrenodoxin. All three proteins together constitute the cholesterol side-chain cleavage complex.
- Hanukoglu I, Rapoport R (1995). “Routes and regulation of NADPH production in steroidogenic mitochondria”. Endocrine Research. 21 (1–2): 231–241. doi:10.3109/07435809509030439. PMID 7588385.
- Hanukoglu I, Gutfinger T, Haniu M, Shively JE (December 1987). “Isolation of a cDNA for adrenodoxin reductase (ferredoxin-NADP+ reductase). Implications for mitochondrial cytochrome P-450 systems”. European Journal of Biochemistry. 169 (3): 449–455. doi:10.1111/j.1432-1033.1987.tb13632.x. PMID 3691502.
- Hanukoglu I, Jefcoate CR (April 1980). “Mitochondrial cytochrome P-450scc. Mechanism of electron transport by adrenodoxin”. The Journal of Biological Chemistry. 255 (7): 3057–3061. doi:10.1016/S0021-9258(19)85851-9. PMID 6766943.
- Hanukoglu I, Spitsberg V, Bumpus JA, Dus KM, Jefcoate CR (May 1981). “Adrenal mitochondrial cytochrome P-450scc. Cholesterol and adrenodoxin interactions at equilibrium and during turnover”. The Journal of Biological Chemistry. 256 (9): 4321–4328. doi:10.1016/S0021-9258(19)69436-6. PMID 7217084.
The involvement of three proteins in cholesterol side-chain cleavage reaction raises the question of whether the three proteins function as a ternary complex as reductase:adrenodoxin:P450. Both spectroscopic studies of adrenodoxin binding to P450scc and kinetic studies in the presence of varying concentrations of adrenodoxin reductase demonstrated that the reductase competes with P450scc for binding to adrenodoxin. These results demonstrated that the formation of a functional ternary complex is not possible. From these studies, it was concluded that the binding sites of adrenodoxin to its reductase and to P450 are overlapping and, as a consequence, adrenodoxin functions as a mobile electron shuttle between reductase and P450. These conclusions have been confirmed by structural analysis of adrenodoxin and P450 complex.
- Hanukoglu I, Jefcoate CR (April 1980). “Mitochondrial cytochrome P-450scc. Mechanism of electron transport by adrenodoxin”. The Journal of Biological Chemistry. 255 (7): 3057–3061. doi:10.1016/S0021-9258(19)85851-9. PMID 6766943.
- Strushkevich N, MacKenzie F, Cherkesova T, Grabovec I, Usanov S, Park HW (June 2011). “Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system”. Proceedings of the National Academy of Sciences of the United States of America. 108 (25): 10139–10143. Bibcode:2011PNAS..10810139S. doi:10.1073/pnas.1019441108. PMC 3121847. PMID 21636783.
The process of electron transfer from NADPH to P450scc is not tightly coupled; that is, during electron transfer from adrenodoxin reductase via adrenodoxin to P450scc, a certain portion of the electrons leak outside of the chain and react with O2, generating superoxide radicals. Steroidogenic cells include a diverse array of antioxidant systems to cope with the radicals generated by the steroidogenic enzymes.
- Hanukoglu I, Rapoport R, Weiner L, Sklan D (September 1993). “Electron leakage from the mitochondrial NADPH-adrenodoxin reductase-adrenodoxin-P450scc (cholesterol side chain cleavage) system”. Archives of Biochemistry and Biophysics. 305 (2): 489–498. doi:10.1006/abbi.1993.1452. PMID 8396893.
- Hanukoglu I (2006). “Antioxidant protective mechanisms against reactive oxygen species (ROS) generated by mitochondrial P450 systems in steroidogenic cells”. Drug Metabolism Reviews. 38 (1–2): 171–196. doi:10.1080/03602530600570040. PMID 16684656. S2CID 10766948.
Regulation
In each steroidogenic cell, the expression of the P450scc system proteins is regulated by the trophic hormonal system specific for the cell type. In adrenal cortex cells from zona fasciculata, the expression of the mRNAs encoding all three P450scc proteins is induced by corticotropin (ACTH). The trophic hormones increase CYP11A1 gene expression through transcription factors such as steroidogenic factor 1 (SF-1), by the α isoform of activating protein 2 (AP-2) in the human, and many others. The production of this enzyme is inhibited notably by the nuclear receptor DAX-1.
- Lavoie HA, King SR (August 2009). “Transcriptional regulation of steroidogenic genes: STARD1, CYP11A1 and HSD3B”. Experimental Biology and Medicine. 234 (8): 880–907. doi:10.3181/0903-MR-97. PMID 19491374. S2CID 5350278.
- Guo IC, Shih MC, Lan HC, Hsu NC, Hu MC, Chung BC (July 2007). “Transcriptional regulation of human CYP11A1 in gonads and adrenals”. Journal of Biomedical Science. 14 (4): 509–515. doi:10.1007/s11373-007-9177-z. PMID 17594537.
- Hanukoglu I, Feuchtwanger R, Hanukoglu A (November 1990). “Mechanism of corticotropin and cAMP induction of mitochondrial cytochrome P450 system enzymes in adrenal cortex cells”. The Journal of Biological Chemistry. 265 (33): 20602–20608. doi:10.1016/S0021-9258(17)30545-8. PMID 2173715.
- Hanukoglu I (December 1992). “Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis”. The Journal of Steroid Biochemistry and Molecular Biology. 43 (8): 779–804. doi:10.1016/0960-0760(92)90307-5. PMID 22217824. S2CID 112729
P450scc is always active, however its activity is limited by the supply of cholesterol in the inner membrane. The supplying of cholesterol to this membrane (from the outer mitochondrial membrane) is, thus, considered the true rate-limiting step in steroid production. This step is mediated primarily by the steroidogenic acute regulatory protein (StAR or STARD1). Upon stimulation of a cell to make steroid, the amount of StAR available to transfer cholesterol to the inner membrane limits how fast the reaction can go (the acute phase). With prolonged (chronic) stimulation, it is thought that cholesterol supply becomes no longer an issue and that the capacity of the system to make steroid (i.e., level of P450scc in the mitochondria) is now more important.
Corticotropin (ACTH) is a hormone that is released from the anterior pituitary in response to stress situations. A study of the steroidogenic capacity of the adrenal cortex in infants with acute respiratory disease demonstrated that indeed during disease state there is a specific increase in the steroidogenic capacity for the synthesis of the glucocorticoid cortisol but not for the mineralocorticoid aldosterone or androgen DHEAS that are secreted from other zones of the adrenal cortex.
- Hanukoglu A, Fried D, Nakash I, Hanukoglu I (November 1995). “Selective increases in adrenal steroidogenic capacity during acute respiratory disease in infants”. European Journal of Endocrinology. 133 (5): 552–556. doi:10.1530/eje.0.1330552. PMID 7581984. S2CID 44439040.
Mutations in the CYP11A1 gene result in a steroid hormone deficiency, causing a minority of cases of the rare and potentially fatal condition lipoid congenital adrenal hyperplasia. Deficiency of CYP11A1 can result in hyperpigmentation, hypoglycemia, and recurrent infections.
- Bhangoo A, Anhalt H, Ten S, King SR (March 2006). “Phenotypic variations in lipoid congenital adrenal hyperplasia”. Pediatric Endocrinology Reviews. 3 (3): 258–271. PMID 16639391.
- al Kandari H, Katsumata N, Alexander S, Rasoul MA (August 2006). “Homozygous mutation of P450 side-chain cleavage enzyme gene (CYP11A1) in 46, XY patient with adrenal insufficiency, complete sex reversal, and agenesis of corpus callosum”. The Journal of Clinical Endocrinology and Metabolism. 91 (8): 2821–2826. doi:10.1210/jc.2005-2230. PMID 16705068.
- Kim CJ, Lin L, Huang N, Quigley CA, AvRuskin TW, Achermann JC, Miller WL (March 2008). “Severe combined adrenal and gonadal deficiency caused by novel mutations in the cholesterol side chain cleavage enzyme, P450scc”. The Journal of Clinical Endocrinology and Metabolism. 93 (3): 696–702. doi:10.1210/jc.2007-2330. PMC 2266942. PMID 18182448.
- FBuonocore F, Achermann JC (2020). “Primary adrenal insufficiency: New genetic causes and their long-term consequences”. Clinical Endocrinology. 92 (1): 11–20. doi:10.1111/cen.14109. PMC 6916405. PMID 31610036.
Cholesterol side-chain cleavage enzyme inhibitors include aminoglutethimide, ketoconazole, and mitotane, among others.
- Becker KL (2001). Principles and Practice of Endocrinology and Metabolism. Lippincott Williams & Wilkins. pp. 735–. ISBN 978-0-7817-1750-2.
- Jameson JL, De Groot LJ (18 May 2010). Endocrinology – E-Book: Adult and Pediatric. Elsevier Health Sciences. pp. 301–302. ISBN 978-1-4557-1126-0.
- Ortiz de Montellano PR (13 March 2015). Cytochrome P450: Structure, Mechanism, and Biochemistry. Springer. pp. 851–879. ISBN 978-3-319-12108-6.
The steroidogenic acute regulatory protein, commonly referred to as StAR (STARD1), is a transport protein that regulates cholesterol transfer within the mitochondria, which is the rate-limiting step in the production of steroid hormones. It is primarily present in steroid-producing cells, including theca cells and luteal cells in the ovary, Leydig cells in the testis and cell types in the adrenal cortex.
StAR is a mitochondrial protein that is rapidly synthesized in response to stimulation of the cell to produce steroid. Hormones that stimulate its production depend on the cell type and include luteinizing hormone (LH), ACTH and angiotensin II. At the cellular level, StAR is synthesized typically in response to activation of the cAMP second messenger system, although other systems can be involved even independently of cAMP. StAR has thus far been found in all tissues that can produce steroids, including the adrenal cortex, the gonads, the brain and the nonhuman placenta. One known exception is the human placenta.
- Wilhelm LP, Wendling C, Védie B, Kobayashi T, Chenard MP, Tomasetto C, Drin G, Alpy F (May 2017). “STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites”. The EMBO Journal. 36 (10): 1412–1433. doi:10.15252/embj.201695917. PMC 5430228. PMID 28377464.
- Alpy F, Stoeckel ME, Dierich A, Escola JM, Wendling C, Chenard MP, Vanier MT, Gruenberg J, Tomasetto C, Rio MC (February 2001). “The steroidogenic acute regulatory protein homolog MLN64, a late endosomal cholesterol-binding protein”. The Journal of Biological Chemistry. 276 (6): 4261–9. doi:10.1074/jbc.M006279200. PMID 11053434.
Substances that suppress StAR activity, like those listed below, can cause endocrine disrupting effects, including altered steroid hormone levels and fertility.
- Alcohol
- DEHP and DBP
- Bis(2-ethylhexyl) phthalate (di-2-ethylhexyl phthalate, diethylhexyl phthalate, diisooctyl phthalate, DEHP; incorrectly — dioctyl phthalate, DIOP)
- Dibutyl phthalate (DBP) is an organic compound which is commonly used as a plasticizer because of its low toxicity and wide liquid range.
- Permethrin and cypermethrin
- Permethrin is a medication and an insecticide.
- Cypermethrin (CP) is a synthetic pyrethroid used as an insecticide in large-scale commercial agricultural applications as well as in consumer products for domestic purposes.
- DES and arsenite
- Diethylstilbestrol (DES), also known as stilbestrol or stilboestrol, is a nonsteroidal estrogen medication
- BPA
- Srivastava VK, Vijayan E, Hiney JK, Dees WL (October 2005). “Effect of ethanol on follicle stimulating hormone-induced steroidogenic acute regulatory protein (StAR) in cultured rat granulosa cells”. Alcohol. 37 (2): 105–11. doi:10.1016/j.alcohol.2006.01.001. PMID 16584974.
- Kariyazono Y, Taura J, Hattori Y, Ishii Y, Narimatsu S, Fujimura M, Takeda T, Yamada H (December 2015). “Effect of in utero exposure to endocrine disruptors on fetal steroidogenesis governed by the pituitary-gonad axis: a study in rats using different ways of administration”. The Journal of Toxicological Sciences. 40 (6): 909–16. doi:10.2131/jts.40.909. PMID 26558472.
- Motohashi M, Wempe MF, Mutou T, Okayama Y, Kansaku N, Takahashi H, Ikegami M, Asari M, Wakui S (2016). “In utero-exposed di(n-butyl) phthalate induce dose dependent, age-related changes of morphology and testosterone-biosynthesis enzymes/associated proteins of Leydig cell mitochondria in rats”. The Journal of Toxicological Sciences. 41 (2): 195–206. doi:10.2131/jts.41.195. PMID 26961603.
- Jin Y, Liu J, Wang L, Chen R, Zhou C, Yang Y, Liu W, Fu Z (2012). “Permethrin exposure during puberty has the potential to enantioselectively induce reproductive toxicity in mice”. Environment International. 42: 144–151. doi:10.1016/j.envint.2011.05.020. ISSN 0160-4120. PMID 21745691.
- Wang H, Wang Q, Zhao X, Liu P, Meng X, Yu T, Ji Y, Zhang H, Zhang C, Zhang Y, Xu D (2009). “Cypermethrin exposure during puberty disrupts testosterone synthesis via downregulating StAR in mouse testes”. Archives of Toxicology. 84 (1): 53–61. doi:10.1007/s00204-009-0479-y. ISSN 0340-5761. PMID 19862501. S2CID 22210562.
- Clark BJ, Cochrum RK (2007). “The Steroidogenic Acute Regulatory Protein as a Target of Endocrine Disruption in Male Reproduction”. Drug Metabolism Reviews. 39 (2–3): 353–370. doi:10.1080/03602530701519151. PMID 17786626. S2CID 26531354.
StAR-independent steroidogenesis
While loss of functional StAR in the human and the mouse catastrophically reduces steroid production, it does not eliminate all of it, indicating the existence of StAR-independent pathways for steroid generation. Aside from the human placenta, these pathways are considered minor for endocrine production. It is unclear what factors catalyze StAR-independent steroidogenesis. Candidates include oxysterols which can be freely converted to steroid and the ubiquitous MLN64.
- Hutson JC (January 2006). “Physiologic interactions between macrophages and Leydig cells”. Exp. Biol. Med. (Maywood). 231 (1): 1–7. doi:10.1177/153537020623100101. PMID 16380639. S2CID 43006988.
StAR related lipid transfer domain containing 3(STARD3) is a protein that in humans is encoded by the STARD3 gene. STARD3 also known as metastatic lymph node 64 protein (MLN64) is a late endosomalintegral membrane protein involved in cholesterol transport. STARD3 creates membrane contact sites between the endoplasmic reticulum (ER) and late endosomes where it moves cholesterol. This gene encodes a member of a subfamily of lipid trafficking proteins that are characterized by a C-terminal steroidogenic acute regulatory domain and an N-terminal metastatic lymph node 64 domain. The encoded protein localizes to the membranes of late endosomes and may be involved in exporting cholesterol. Alternative splicing results in multiple transcript variants.[provided by RefSeq, Oct 2009]. STARD3 is involved in cholesterol transport from the ER to late endosomes where the protein is anchored. It forms a complex with fellow late endosomal protein STARD3 N-terminal-like protein (STARD3NL) also known as MLN64 N-terminal homologue (MENTHO) and ER VAMP-associated proteins (VAP proteins) A and B (VAP-A, VAP-B) to tether the two organelles together. For STARD3, this interaction is regulated by phosphorylation of a serine in its FFAT motif. The closest homolog to STARD3 is the steroidogenic acute regulatory protein (StAR/StarD1), which initiates the production of steroids by moving cholesterol inside the mitochondrion. Thus, MLN64 is also proposed to move cholesterol inside the mitochondria under certain conditions to initiate StAR-independent steroidogenesis, such as in the human placenta which lacks StAR yet produces steroids. This functional role is supported by evidence that MLN64 expression can stimulate steroid production in a model cell system.
- “Entrez Gene: StAR related lipid transfer domain containing 3”. Retrieved 2018-08-07.
- Alpy F, Tomasetto C (June 2006). “MLN64 and MENTHO, two mediators of endosomal cholesterol transport”. Biochemical Society Transactions. 34 (Pt 3): 343–5. doi:10.1042/BST0340343. PMID 16709157.
- Wilhelm LP, Wendling C, Védie B, Kobayashi T, Chenard MP, Tomasetto C, Drin G, Alpy F (May 2017). “STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites”. The EMBO Journal. 36 (10): 1412–1433. doi:10.15252/embj.201695917. PMC 5430228. PMID 28377464.
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio MC, Levine TP, Tomasetto C (December 2013). “STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER”. Journal of Cell Science. 126 (Pt 23): 5500–12. doi:10.1242/jcs.139295. PMID 24105263.
- Wilhelm LP, Wendling C, Védie B, Kobayashi T, Chenard MP, Tomasetto C, Drin G, Alpy F (May 2017). “STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites”. The EMBO Journal. 36 (10): 1412–1433. doi:10.15252/embj.201695917. PMC 5430228. PMID 28377464.
- Alpy F, Stoeckel ME, Dierich A, Escola JM, Wendling C, Chenard MP, Vanier MT, Gruenberg J, Tomasetto C, Rio MC (February 2001). “The steroidogenic acute regulatory protein homolog MLN64, a late endosomal cholesterol-binding protein”. The Journal of Biological Chemistry. 276 (6): 4261–9. doi:10.1074/jbc.M006279200. PMID 11053434.
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio MC, Levine TP, Tomasetto C (December 1, 2013). “STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER” (PDF). J Cell Sci. 126 (23): 5500–5512. doi:10.1242/jcs.139295. PMID 24105263. S2CID 7245863.
- Di Mattia, Thomas; Martinet, Arthur; Ikhlef, Souade; McEwen, Alastair G; Nominé, Yves; Wendling, Corinne; Poussin-Courmontagne, Pierre; Voilquin, Laetitia; Eberling, Pascal; Ruffenach, Frank; Cavarelli, Jean; Slee, John; Levine, Timothy P; Drin, Guillaume; Tomasetto, Catherine; Alpy, Fabien (December 1, 2020). “FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts”. The EMBO Journal. 39 (23): e104369. doi:10.15252/embj.2019104369. ISSN 0261-4189. PMC 7705450. PMID 33124732.
- Watari H, Arakane F, Moog-Lutz C, Kallen CB, Tomasetto C, Gerton GL, Rio MC, Baker ME, Strauss JF (August 1997). “MLN64 contains a domain with homology to the steroidogenic acute regulatory protein (StAR) that stimulates steroidogenesis”. Proceedings of the National Academy of Sciences of the United States of America. 94 (16): 8462–7. Bibcode:1997PNAS…94.8462W. doi:10.1073/pnas.94.16.8462. PMC 22957. PMID 9237999.
One study indicates that STARD3 (MLN64) also specifically binds lutein in the retina.
- Li B, Vachali P, Frederick JM, Bernstein PS (April 2011). “Identification of StARD3 as a lutein-binding protein in the macula of the primate retina”. Biochemistry. 50 (13): 2541–9. doi:10.1021/bi101906y. PMC 3070171. PMID 21322544.
STARD3 is a multi-domain protein composed of a N-terminal MENTAL (MLN64 N-terminal) domain, a central phospho-FFAT motif (two phenylalanines in an acidic tract), and a C-terminal StAR-related transfer domain (START) lipid transport domain. The MENTAL domain of STARD3 is similar to the protein STARD3 N-terminal like protein (STARD3NL) also known as MLN64 N-terminal homologue (MENTHO). This domain is composed of 4 transmembrane helices which anchor the protein in the limiting membrane of late endosomes. This domain binds cholesterol and associates with the same domain in STARD3NL. The phospho-FFAT motif is a short protein sequence motif which binds to the ER proteins VAP-A, VAP-B and MOSPD2 proteins after phosphorylation. The START domain of STARD3 is homologous to the StAR protein. X-ray crystallography of the C-terminus indicates that this domain forms a pocket that can bind cholesterol. This places STARD3 within the StarD1/D3 subfamily of START domain-containing proteins.
- Alpy F, Wendling C, Rio MC, Tomasetto C (December 2002). “MENTHO, a MLN64 homologue devoid of the START domain”. The Journal of Biological Chemistry. 277 (52): 50780–7. doi:10.1074/jbc.M208290200. PMID 12393907.
- Alpy F, Latchumanan VK, Kedinger V, Janoshazi A, Thiele C, Wendling C, Rio MC, Tomasetto C (May 2005). “Functional characterization of the MENTAL domain”. The Journal of Biological Chemistry. 280 (18): 17945–52. doi:10.1074/jbc.M500723200. PMID 15718238.
- Di Mattia, Thomas; Martinet, Arthur; Ikhlef, Souade; McEwen, Alastair G; Nominé, Yves; Wendling, Corinne; Poussin-Courmontagne, Pierre; Voilquin, Laetitia; Eberling, Pascal; Ruffenach, Frank; Cavarelli, Jean; Slee, John; Levine, Timothy P; Drin, Guillaume; Tomasetto, Catherine; Alpy, Fabien (December 1, 2020). “FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts”. The EMBO Journal. 39 (23): e104369. doi:10.15252/embj.2019104369. ISSN 0261-4189. PMC 7705450. PMID 33124732.
- Tsujishita Y, Hurley JH (May 2000). “Structure and lipid transport mechanism of a StAR-related domain”. Nature Structural Biology. 7 (5): 408–14. doi:10.1038/75192. PMID 10802740. S2CID 10806665.
Tissue distribution (MLN64)
STARD3 is expressed in all tissues in the body at various levels. In the brain, MLN64 is detectable in many but not all cells. Many malignant tumors highly express STARD3 as a result of its gene being part of a Her2/erbB2-containing gene locus that is amplified.
- King SR, Smith AG, Alpy F, Tomasetto C, Ginsberg SD, Lamb DJ (2006). “Characterization of the putative cholesterol transport protein metastatic lymph node 64 in the brain”. Neuroscience. 139 (3): 1031–8. doi:10.1016/j.neuroscience.2006.01.063. PMID 16549269. S2CID 33113555.
New roles (StAR)
Recent findings suggest that StAR may also traffic cholesterol to a second mitochondrial enzyme, sterol 27-hydroxylase. This enzyme converts cholesterol to 27-hydroxycholesterol. In this way it may be important for the first step in one of the two pathways for the production of bile acids by the liver (the alternative pathway). Evidence also shows that the presence of StAR in a type of immune cell, the macrophage, where it can stimulate the production of 27-hydroxycholesterol. In this case, 27-hydroxycholesterol may by itself be helpful against the production of inflammatory factors associated with cardiovascular disease. It is important to note that no study has yet found a link between the loss of StAR and problems in bile acid production or increased risk for cardiovascular disease. Recently StAR was found to be expressed in cardiac fibroblasts in response to ischemic injury due to myocardial infarction. In these cells it has no apparent de novo steroidogenic activity, as evidenced by the lack of the key steroidogenic enzymes cytochrome P450 side chain cleavage (CYP11A1) and 3 beta hydroxysteroid dehydrogenase (3βHSD). StAR was found to have an anti-apoptotic effect on the fibroblasts, which may allow them to survive the initial stress of the infarct, differentiate and function in tissue repair at the infarction site.
- Hall EA, Ren S, Hylemon PB, Rodriguez-Agudo D, Redford K, Marques D, Kang D, Gil G, Pandak WM (April 2005). “Detection of the steroidogenic acute regulatory protein, StAR, in human liver cells”. Biochim Biophys Acta. 1733 (2–3): 111–119. doi:10.1016/j.bbalip.2005.01.004. PMID 15863358.
- Ma Y, Ren S, Pandak WM, Li X, Ning Y, Lu C, Zhao F, Yin L (December 2007). “The effects of inflammatory cytokines on steroidogenic acute regulatory protein expression in macrophages”. Inflamm Res. 56 (12): 495–501. doi:10.1007/s00011-007-6133-3. PMID 18210233. S2CID 21308251.
- Taylor JM, Borthwick F, Bartholomew C, Graham A (June 2010). “Overexpression of steroidogenic acute regulatory protein increases macrophage cholesterol efflux to apolipoprotein AI”. Cardiovasc Res. 86 (3): 526–534. doi:10.1093/cvr/cvq015. PMID 20083572.
- Anuka E, Yivgi-Ohana N, Eimerl S, Garfinkel B, Melamed-Book N, Chepurkol E, Aravot D, Zinman T, Shainberg A, Hochhauser E, Orly J (September 2013). “Infarct-Induced Steroidogenic Acute Regulatory Protein: A Survival Role in Cardiac Fibroblasts”. Mol Endocrinol. 27 (9): 1502–1517. doi:10.1210/me.2013-1006. PMC 5415234. PMID 23831818.
SF-1 has been identified as a transcriptional regulator for an array of different genes related to sex determination and differentiation, reproduction, and metabolism via binding to their promoters. For example, SF-1 controls expression of Amh gene in Sertoli cells, whereby the presence or absence of the gene product affects development of Müllerian structures. Increased AMH protein levels leads to regression of such structures. Leydig cells express SF-1 to regulate transcription of steroidogenesis and testosterone biosynthesis genes causing virilization in males.
- Parker KL, Schimmer BP (June 1997). “Steroidogenic factor 1: a key determinant of endocrine development and function”. Endocrine Reviews. 18 (3): 361–77. doi:10.1210/edrv.18.3.0301. PMID 9183568.
Target genes
Steroidogenic cells
First identified as a regulator of steroid hydroxylases within adrenocortical cells, studies aimed to define localization and expression of SF-1 have since revealed enzyme activity within other steroidogenic cells.
- Parker KL, Schimmer BP (June 1997). “Steroidogenic factor 1: a key determinant of endocrine development and function”. Endocrine Reviews. 18 (3): 361–77. doi:10.1210/edrv.18.3.0301. PMID 9183568.
| species | Gene | Cell/Tissue |
|---|---|---|
| rat | P450scc | granulosa cells |
| mouse | P450scc | Y1 adrenocortical cells |
| bovine | Oxytocin | ovary |
| mouse | StAR | MA-10 Leydig cells |
Sertoli cells
The Müllerian inhibiting substance (MIS or AMH) gene within Sertoli cells contains a conserved motif identical to the optimal binding sequence for SF-1. Gel mobility shift experiments and use of SF-1-specific polyclonal antibodies established binding complexes of SF-1 to MIS, however, other studies suggest the MIS promoter is repressed and not activated by SF-1 binding.
- Shen WH, Moore CC, Ikeda Y, Parker KL, Ingraham HA (June 1994). “Nuclear receptor steroidogenic factor 1 regulates the müllerian inhibiting substance gene: a link to the sex determination cascade”. Cell. 77 (5): 651–61. doi:10.1016/0092-8674(94)90050-7. PMID 8205615. S2CID 13364008.
Gonadotropes
Gonadotrope-specific element, or GSE, in the promoter of the gene encoding α-subunit of glycoproteins (α-GSU) resembles the SF-1 binding sires. Studies have implicated SF-1 as an upstream regulator of a collection of genes required for gonadotrope function via GSE.
- Ingraham HA, Lala DS, Ikeda Y, Luo X, Shen WH, Nachtigal MW, Abbud R, Nilson JH, Parker KL (October 1994). “The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis”. Genes & Development. 8 (19): 2302–12. doi:10.1101/gad.8.19.2302. PMID 7958897.
VMH
SF-1 knockout mice displayed profound defects in the VMH suggesting potential target genes at the site. Target genes have yet to be identified due to difficulties in studying gene expression in neurons.
SF-1 gene knockout
Several approaches used targeted gene disruption in mouse embryonic stem cells with the aim of identifying potential target genes of SF-1. The different targeting strategies include disruption to exons encoding for the zinc finger motif, disruption of a 3’-exon and targeted mutation of the initiator methionine. The corresponding observed phenotypic effects on endocrine development and function were found to be quite similar.
- Parker KL, Schimmer BP (June 1997). “Steroidogenic factor 1: a key determinant of endocrine development and function”. Endocrine Reviews. 18 (3): 361–77. doi:10.1210/edrv.18.3.0301. PMID 9183568.
Sf-1 knockout mice displayed diminished corticosterone levels while maintaining elevated ACTH levels. Observed morphological changes and DNA fragmentation was consistent with apoptosis and structural regression resulting in the death of all mice within 8 days after birth.
- Luo X, Ikeda Y, Schlosser DA, Parker KL (September 1995). “Steroidogenic factor 1 is the essential transcript of the mouse Ftz-F1 gene”. Molecular Endocrinology. 9 (9): 1233–9. doi:10.1210/mend.9.9.7491115. PMID 7491115.
Sf-1 function was determined to be necessary for development of primary steroidogenic tissue as evidenced by complete lack of adrenal and gonadal glands in the knockout. Male to female sex reversal of genitalia was also observed.
- Luo X, Ikeda Y, Parker KL (May 1994). “A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation”. Cell. 77 (4): 481–90. doi:10.1016/0092-8674(94)90211-9. PMID 8187173. S2CID 28194376.
Clinical significance
Mutations in NR5A1 can produce intersex genitals, absence of puberty, and infertility. It is one cause of arrest of ovarian function in women <40 years of age, which occurs in 1% of all women.
Adrenal and gonadal failure
Two SF-1 variants associated with primary adrenal failure and complete gonadal dysgenesis have been reported as caused by NR5A1 mutations. One reported case was found to have de novo heterozygous p.G35E change to the P-box domain. The affected region allows for DNA binding specificity through interactions with regulatory response elements of target genes. This p.G35E change may have a mild competitive or dominant negative effect on transactivation resulting in severe gonadal defects and adrenal dysfunction. Similarly, homozygous p.R92Q change within the A-box interfered with monomeric binding stability and reduced functional activity. This change requires mutations to both allele to display phenotypic effects as heterozygous carriers showed normal adrenal function.
- Ferraz-de-Souza B, Lin L, Achermann JC (April 2011). “Steroidogenic factor-1 (SF-1, NR5A1) and human disease”. Molecular and Cellular Endocrinology. 336 (1–2): 198–205. doi:10.1016/j.mce.2010.11.006. PMC 3057017. PMID 21078366.
Missense, in-frame and frameshift mutations of NR5A1 have been found in families with 46,XY disorders of sex development, 46,XX gonadal dysgenesis and 46,XX primary ovarian insufficiency. 46,XY individuals may have ambiguous or female genitals. Individuals of either karyotype may not enter puberty, although expression of the phenotype, penetrance, fertility, and modes of inheritance can vary. Some mutations are dominant, some are recessive.
- Lourenço D, Brauner R, Lin L, De Perdigo A, Weryha G, Muresan M, Boudjenah R, Guerra-Junior G, Maciel-Guerra AT, Achermann JC, McElreavey K, Bashamboo A (March 2009). “Mutations in NR5A1 associated with ovarian insufficiency”. The New England Journal of Medicine. 360 (12): 1200–10. doi:10.1056/NEJMoa0806228. PMC 2778147. PMID 19246354.
46, XY disorders of sex development
Heterozygous NR5A1 changes are emerging as a frequent contributor in 46, XY complete gonadal dysgenesis. In affected individuals, sexual development does not match their chromosomal makeup. Males, despite having 46, XY karyotype, develop female external genitalia, as well as uterus and fallopian tubes, along with gonadal defects rendering them nonfunctional. NR5A1 mutations have also been linked to partial gonadal dysgenesis, whereby affected individuals have ambiguous genitalia, urogenital sinus, absent or rudimentary Müllerian structures, and other abnormalities.
- Ferraz-de-Souza B, Lin L, Achermann JC (April 2011). “Steroidogenic factor-1 (SF-1, NR5A1) and human disease”. Molecular and Cellular Endocrinology. 336 (1–2): 198–205. doi:10.1016/j.mce.2010.11.006. PMC 3057017. PMID 21078366.
- Reference, Genetics Home. “Swyer syndrome”. Genetics Home Reference. Retrieved 2017-11-30.
Typically, these genetic changes are frameshift, nonsense, or missense mutations that alter DNA-binding and gene transcription. While many are de novo, one-third of cases have been maternally inherited in a similar manner as X-linked inheritance. Furthermore, one report of homozygous missense mutation p.D293N within the ligand-binding domain of SF-1 revealed autosomal recessive inheritance was also possible.
- Lourenço D, Brauner R, Lin L, De Perdigo A, Weryha G, Muresan M, Boudjenah R, Guerra-Junior G, Maciel-Guerra AT, Achermann JC, McElreavey K, Bashamboo A (March 2009). “Mutations in NR5A1 associated with ovarian insufficiency”. The New England Journal of Medicine. 360 (12): 1200–10. doi:10.1056/NEJMoa0806228. PMC 2778147. PMID 19246354.
Infertility
Analysis of NR5A1 in men with non-obstructive male factor infertility found those with gene changes had more severe forms of infertility and lower testosterone levels. These changes affected the hinge region of SF-1. It is important to note further studies are required to establish the relationship between SF-1 changes and infertility.
- Bashamboo A, Ferraz-de-Souza B, Lourenço D, Lin L, Sebire NJ, Montjean D, Bignon-Topalovic J, Mandelbaum J, Siffroi JP, Christin-Maitre S, Radhakrishna U, Rouba H, Ravel C, Seeler J, Achermann JC, McElreavey K (October 2010). “Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1”. American Journal of Human Genetics. 87 (4): 505–12. doi:10.1016/j.ajhg.2010.09.009. PMC 2948805. PMID 20887963.
Additional interactions
SF-1 has also been shown to interact with:
- Beta-catenin[24][25]
- DAX1[26][27]
- NRIP1[28][29]
- SOX9[30]
- TRERF1.[31]
References
- GRCh38: Ensembl release 89: ENSG00000136931 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000026751 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- Reference, Genetics Home. “NR5A1 gene”. Genetics Home Reference. Retrieved 2017-11-30.
- Parker KL, Schimmer BP (June 1997). “Steroidogenic factor 1: a key determinant of endocrine development and function”. Endocrine Reviews. 18 (3): 361–77. doi:10.1210/edrv.18.3.0301. PMID 9183568.
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM (December 1995). “The nuclear receptor superfamily: the second decade”. Cell. 83 (6): 835–9. doi:10.1016/0092-8674(95)90199-x. PMC 6159888. PMID 8521507.
- Honda S, Morohashi K, Nomura M, Takeya H, Kitajima M, Omura T (April 1993). “Ad4BP regulating steroidogenic P-450 gene is a member of steroid hormone receptor superfamily”. The Journal of Biological Chemistry. 268 (10): 7494–502. doi:10.1016/S0021-9258(18)53202-6. PMID 8463279.
- Ikeda Y, Lala DS, Luo X, Kim E, Moisan MP, Parker KL (July 1993). “Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression”. Molecular Endocrinology. 7 (7): 852–60. doi:10.1210/mend.7.7.8413309. PMID 8413309.
- Morohashi K, Iida H, Nomura M, Hatano O, Honda S, Tsukiyama T, Niwa O, Hara T, Takakusu A, Shibata Y (May 1994). “Functional difference between Ad4BP and ELP, and their distributions in steroidogenic tissues”. Molecular Endocrinology. 8 (5): 643–53. doi:10.1210/mend.8.5.8058072. PMID 8058072.
- Takayama K, Sasano H, Fukaya T, Morohashi K, Suzuki T, Tamura M, Costa MJ, Yajima A (September 1995). “Immunohistochemical localization of Ad4-binding protein with correlation to steroidogenic enzyme expression in cycling human ovaries and sex cord stromal tumors”. The Journal of Clinical Endocrinology and Metabolism. 80 (9): 2815–21. doi:10.1210/jcem.80.9.7673429. PMID 7673429.
- “Male Development of Chromosomally Female Mice Transgenic for Sry gene” (1991), by Peter Koopman, et al. | The Embryo Project Encyclopedia”. embryo.asu.edu. Retrieved 2017-11-30.
- Ninomiya Y, Okada M, Kotomura N, Suzuki K, Tsukiyama T, Niwa O (1995). “Genomic organization and isoforms of the mouse ELP gene”. Journal of Biochemistry. 118 (2): 380–9. doi:10.1093/oxfordjournals.jbchem.a124918. PMID 8543574.
- Lewis AE, Rusten M, Hoivik EA, Vikse EL, Hansson ML, Wallberg AE, Bakke M (January 2008). “Phosphorylation of steroidogenic factor 1 is mediated by cyclin-dependent kinase 7”. Molecular Endocrinology. 22 (1): 91–104. doi:10.1210/me.2006-0478. PMC 5419630. PMID 17901130.
- Jameson JL (December 2004). “Of mice and men: The tale of steroidogenic factor-1”. The Journal of Clinical Endocrinology and Metabolism. 89 (12): 5927–9. doi:10.1210/jc.2004-2047. PMID 15579738.
- Shen WH, Moore CC, Ikeda Y, Parker KL, Ingraham HA (June 1994). “Nuclear receptor steroidogenic factor 1 regulates the müllerian inhibiting substance gene: a link to the sex determination cascade”. Cell. 77 (5): 651–61. doi:10.1016/0092-8674(94)90050-7. PMID 8205615. S2CID 13364008.
- Ingraham HA, Lala DS, Ikeda Y, Luo X, Shen WH, Nachtigal MW, Abbud R, Nilson JH, Parker KL (October 1994). “The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis”. Genes & Development. 8 (19): 2302–12. doi:10.1101/gad.8.19.2302. PMID 7958897.
- Luo X, Ikeda Y, Schlosser DA, Parker KL (September 1995). “Steroidogenic factor 1 is the essential transcript of the mouse Ftz-F1 gene”. Molecular Endocrinology. 9 (9): 1233–9. doi:10.1210/mend.9.9.7491115. PMID 7491115.
- Luo X, Ikeda Y, Parker KL (May 1994). “A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation”. Cell. 77 (4): 481–90. doi:10.1016/0092-8674(94)90211-9. PMID 8187173. S2CID 28194376.
- Ferraz-de-Souza B, Lin L, Achermann JC (April 2011). “Steroidogenic factor-1 (SF-1, NR5A1) and human disease”. Molecular and Cellular Endocrinology. 336 (1–2): 198–205. doi:10.1016/j.mce.2010.11.006. PMC 3057017. PMID 21078366.
- Lourenço D, Brauner R, Lin L, De Perdigo A, Weryha G, Muresan M, Boudjenah R, Guerra-Junior G, Maciel-Guerra AT, Achermann JC, McElreavey K, Bashamboo A (March 2009). “Mutations in NR5A1 associated with ovarian insufficiency”. The New England Journal of Medicine. 360 (12): 1200–10. doi:10.1056/NEJMoa0806228. PMC 2778147. PMID 19246354.
- Reference, Genetics Home. “Swyer syndrome”. Genetics Home Reference. Retrieved 2017-11-30.
- Bashamboo A, Ferraz-de-Souza B, Lourenço D, Lin L, Sebire NJ, Montjean D, Bignon-Topalovic J, Mandelbaum J, Siffroi JP, Christin-Maitre S, Radhakrishna U, Rouba H, Ravel C, Seeler J, Achermann JC, McElreavey K (October 2010). “Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1”. American Journal of Human Genetics. 87 (4): 505–12. doi:10.1016/j.ajhg.2010.09.009. PMC 2948805. PMID 20887963.
- Kennell JA, O’Leary EE, Gummow BM, Hammer GD, MacDougald OA (August 2003). “T-cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with beta-catenin to coactivate C/EBPalpha and steroidogenic factor 1 transcription factors”. Molecular and Cellular Biology. 23 (15): 5366–75. doi:10.1128/MCB.23.15.5366-5375.2003. PMC 165725. PMID 12861022.
- Mizusaki H, Kawabe K, Mukai T, Ariyoshi E, Kasahara M, Yoshioka H, Swain A, Morohashi K (April 2003). “Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) gene transcription is regulated by wnt4 in the female developing gonad”. Molecular Endocrinology. 17 (4): 507–19. doi:10.1210/me.2002-0362. PMID 12554773.
- Lopez D, Shea-Eaton W, Sanchez MD, McLean MP (December 2001). “DAX-1 represses the high-density lipoprotein receptor through interaction with positive regulators sterol regulatory element-binding protein-1a and steroidogenic factor-1”. Endocrinology. 142 (12): 5097–106. doi:10.1210/endo.142.12.8523. PMID 11713202.
- Sugawara T, Saito M, Fujimoto S (August 2000). “Sp1 and SF-1 interact and cooperate in the regulation of human steroidogenic acute regulatory protein gene expression”. Endocrinology. 141 (8): 2895–903. doi:10.1210/endo.141.8.7602. PMID 10919277. S2CID 20567318.
- Mellgren G, Børud B, Hoang T, Yri OE, Fladeby C, Lien EA, Lund J (May 2003). “Characterization of receptor-interacting protein RIP140 in the regulation of SF-1 responsive target genes”. Molecular and Cellular Endocrinology. 203 (1–2): 91–103. doi:10.1016/S0303-7207(03)00097-2. PMID 12782406. S2CID 733221.
- Sugawara T, Abe S, Sakuragi N, Fujimoto Y, Nomura E, Fujieda K, Saito M, Fujimoto S (August 2001). “RIP 140 modulates transcription of the steroidogenic acute regulatory protein gene through interactions with both SF-1 and DAX-1”. Endocrinology. 142 (8): 3570–7. doi:10.1210/endo.142.8.8309. PMID 11459805.
- De Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Sudbeck P, Scherer G, Poulat F, Berta P (November 1998). “Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene”. Molecular and Cellular Biology. 18 (11): 6653–65. doi:10.1128/mcb.18.11.6653. PMC 109250. PMID 9774680.
- Gizard F, Lavallee B, DeWitte F, Teissier E, Staels B, Hum DW (October 2002). “The transcriptional regulating protein of 132 kDa (TReP-132) enhances P450scc gene transcription through interaction with steroidogenic factor-1 in human adrenal cells”. The Journal of Biological Chemistry. 277 (42): 39144–55. doi:10.1074/jbc.M205786200. PMID 12101186.
- Kurrasch DM, Cheung CC, Lee FY, Tran PV, Hata K, Ingraham HA (December 2007). “The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning”. The Journal of Neuroscience. 27 (50): 13624–34. doi:10.1523/JNEUROSCI.2858-07.2007. PMC 6673626. PMID 18077674.
- Gold RM (November 1973). “Hypothalamic obesity: the myth of the ventromedial nucleus”. Science. 182 (4111): 488–90. Bibcode:1973Sci…182..488G. doi:10.1126/science.182.4111.488. PMID 4795550. S2CID 3011420.
- Balagura S, Devenport LD (June 1970). “Feeding patterns of normal and ventromedial hypothalamic lesioned male and female rats”. Journal of Comparative and Physiological Psychology. 71 (3): 357–64. doi:10.1037/h0029118. PMID 5480868.
- Becker EE, Kissileff HR (February 1974). “Inhibitory controls of feeding by the ventromedial hypothalamus”. The American Journal of Physiology. 226 (2): 383–96. doi:10.1152/ajplegacy.1974.226.2.383. PMID 4811195.
- Berthoud HR, Jeanrenaud B (September 1979). “Changes of insulinemia, glycemia and feeding behavior induced by VMH-procainization in the rat”. Brain Research. 174 (1): 184–7. doi:10.1016/0006-8993(79)90816-3. PMID 487120. S2CID 39015121.
- Brooks CM, Lockwood RA, Wiggins ML (December 1946). “A study of the effect of hypothalamic lesions on the eating habits of the albino rat”. The American Journal of Physiology. 147 (4): 735–41. doi:10.1152/ajplegacy.1946.147.4.735. PMID 20277066.
- Epstein AN (December 1960). “Reciprocal changes in feeding behavior produced by intrahypothalamic chemical injections”. The American Journal of Physiology. 199 (6): 969–74. doi:10.1152/ajplegacy.1960.199.6.969. PMID 13697000.
- Larkin RP (November 1975). “Effect of ventromedial hypothalamic procaine injections on feeding, lever pressing, and other behavior in rats”. Journal of Comparative and Physiological Psychology. 89 (9): 1100–8. doi:10.1037/h0077192. PMID 1202103.
- Maes H (June 1980). “Time course of feeding induced by pentobarbital-injections into the rat’s VMH”. Physiology & Behavior. 24 (6): 1107–14. doi:10.1016/0031-9384(80)90055-4. PMID 7413790. S2CID 43051882.
- King BM (February 2006). “The rise, fall, and resurrection of the ventromedial hypothalamus in the regulation of feeding behavior and body weight”. Physiology & Behavior. 87 (2): 221–44. doi:10.1016/j.physbeh.2005.10.007. PMID 16412483. S2CID 40880350.
- Satoh N, Ogawa Y, Katsuura G, Tsuji T, Masuzaki H, Hiraoka J, Okazaki T, Tamaki M, Hayase M, Yoshimasa Y, Nishi S, Hosoda K, Nakao K (March 1997). “Pathophysiological significance of the obese gene product, leptin, in ventromedial hypothalamus (VMH)-lesioned rats: evidence for loss of its satiety effect in VMH-lesioned rats”. Endocrinology. 138 (3): 947–54. doi:10.1210/endo.138.3.4989. PMID 9048594.
- Hetherington AW, Ranson SW (June 1942). “The relation of various hypothalamic lesions to adiposity in the rat” (PDF). Journal of Comparative Neurology. 76 (3): 475–99. doi:10.1002/cne.900760308. S2CID 85715802.
- Jamshidi N, Taylor DA (November 2001). “Anandamide administration into the ventromedial hypothalamus stimulates appetite in rats”. British Journal of Pharmacology. 134 (6): 1151–4. doi:10.1038/sj.bjp.0704379. PMC 1573067. PMID 11704633.
- Panksepp J, Siviy S, Normansell L (1984). “The psychobiology of play: theoretical and methodological perspectives”. Neuroscience and Biobehavioral Reviews. 8 (4): 465–92. doi:10.1016/0149-7634(84)90005-8. PMID 6392950. S2CID 26810046.
- Flanagan-Cato LM, Lee BJ, Calizo LH (June 2006). “Co-localization of midbrain projections, progestin receptors, and mating-induced fos in the hypothalamic ventromedial nucleus of the female rat”. Hormones and Behavior. 50 (1): 52–60. doi:10.1016/j.yhbeh.2006.01.012. PMID 16546183. S2CID 36201218.
- Harding SM, McGinnis MY (October 2005). “Microlesions of the ventromedial nucleus of the hypothalamus: effects on sociosexual behaviors in male rats”. Behavioral Neuroscience. 119 (5): 1227–34. doi:10.1037/0735-7044.119.5.1227. PMID 16300430.
- Kammel LG, Correa SM (January 2020). “Selective sexual differentiation of neurone populations may contribute to sex-specific outputs of the ventromedial nucleus of the hypothalamus”. Journal of Neuroendocrinology. 32 (1): e12801. doi:10.1111/jne.12801. PMC 6982598. PMID 31605642.
- Kow LM, Pfaff DW (May 1998). “Mapping of neural and signal transduction pathways for lordosis in the search for estrogen actions on the central nervous system”. Behavioural Brain Research. 92 (2): 169–80. doi:10.1016/S0166-4328(97)00189-7. PMID 9638959. S2CID 28276218.
- Christensen LW, Nance DM, Gorski RA (1977). “Effects of hypothalamic and preoptic lesions on reproductive behavior in male rats”. Brain Research Bulletin. 2 (2): 137–41. doi:10.1016/0361-9230(77)90010-7. PMID 880486. S2CID 4700161.
- Pfaff DW, Sakuma Y (March 1979). “Facilitation of the lordosis reflex of female rats from the ventromedial nucleus of the hypothalamus”. The Journal of Physiology. 288: 189–202. doi:10.1113/jphysiol.1979.sp012690. PMC 1281421. PMID 469715.
- Matsumoto T, Yamanouchi K (September 2000). “Acceleration of mounting behaviors in female rats by ibotenic acid lesions in the ventromedial hypothalamic nucleus”. Neuroscience Letters. 291 (3): 143–6. doi:10.1016/S0304-3940(00)01388-4. PMID 10984627. S2CID 10334038.
- Correa SM, Newstrom DW, Warne JP, Flandin P, Cheung CC, Lin-Moore AT, Pierce AA, Xu AW, Rubenstein JL, Ingraham HA (January 2015). “An Estrogen-Responsive Module in the Ventromedial Hypothalamus Selectively Drives Sex-Specific Activity in Females”. Cell Reports. 10 (1): 62–74. doi:10.1016/j.celrep.2014.12.011. PMC 4324838. PMID 25543145.
- van Veen JE, Kammel LG, Bunda PC, Shum M, Reid MS, Massa MG, Arneson D, Park JW, Zhang Z, Joseph AM, Hrncir H, Liesa M, Arnold AP, Yang X, Correa SM (April 2020). “Hypothalamic estrogen receptor alpha establishes a sexually dimorphic regulatory node of energy expenditure”. Nature Metabolism. 2 (4): 351–63. doi:10.1038/s42255-020-0189-6. PMC 7202561. PMID 32377634.
- Kadiyala SB, Papandrea D, Tuz K, Anderson TM, Jayakumar S, Herron BJ, Ferland RJ (January 2015). “Spatiotemporal differences in the c-fos pathway between C57BL/6J and DBA/2J mice following flurothyl-induced seizures: A dissociation of hippocampal Fos from seizure activity”. Epilepsy Research. 109: 183–96. doi:10.1016/j.eplepsyres.2014.11.009. PMC 4272448. PMID 25524858.
- Kadiyala SB, Ferland RJ (March 2017). “Dissociation of spontaneous seizures and brainstem seizure thresholds in mice exposed to eight flurothyl-induced generalized seizures”. Epilepsia Open. 2 (1): 48–58. doi:10.1002/epi4.12031. PMC 5560332. PMID 28825051.
- Ferland RJ, Applegate CD (November 1998). “The role of the ventromedial nucleus of the hypothalamus in epileptogenesis”. NeuroReport. 9 (16): 3623–9. doi:10.1097/00001756-199811160-00013. PMID 9858370. S2CID 29713035.
- Rieber, Inge; Sigusch, Volkmar (1979). “Psychosurgery on sex offenders and sexual ?deviants? in West Germany”. Archives of Sexual Behavior. 8 (6): 523–527. doi:10.1007/BF01541419. PMID 391177. S2CID 41463669.
- Kallen CB, Billheimer JT, Summers SA, Stayrook SE, Lewis M, Strauss III JF (October 1998). “Steroidogenic acute regulatory protein (StAR) is a sterol transfer protein”. J. Biol. Chem. 273 (41): 26285–8. doi:10.1074/jbc.273.41.26285. PMID 9756854.
- Bose HS, Whittal RM, Baldwin MA, Miller WL (June 1999). “The active form of the steroidogenic acute regulatory protein, StAR, appears to be a molten globule”. Proc. Natl. Acad. Sci. U.S.A. 96 (13): 7250–5. Bibcode:1999PNAS…96.7250B. doi:10.1073/pnas.96.13.7250. PMC 22068. PMID 10377400.
- Roostaee A, Barbar E, Lehoux JG, Lavigne P (June 2008). “Cholesterol binding is a prerequisite for the activity of the steroidogenic acute regulatory protein (StAR)”. Biochem. J. 412 (3): 553–62. doi:10.1042/BJ20071264. PMID 18341481.
- Christenson LK, Strauss III JF (2001). “Steroidogenic acute regulatory protein: an update on its regulation and mechanism of action”. Arch. Med. Res. 32 (6): 576–86. doi:10.1016/S0188-4409(01)00338-1. PMID 11750733.
- Williams SV, Platt FM, Hurst CD, Aveyard JS, Taylor CF, Pole JC, Garcia MJ, Knowles MA (2010). “High-resolution analysis of genomic alteration on chromosome arm 8p in urothelial carcinoma”. Genes, Chromosomes and Cancer. 49 (7): 642–659. doi:10.1002/gcc.20775. PMID 20461757. S2CID 29971371.
- Arakane F, King SR, Du Y, Kallen CB, Walsh LP, Watari H, Stocco DM, Strauss JF (December 1997). “Phosphorylation of steroidogenic acute regulatory protein (StAR) modulates its steroidogenic activity”. J. Biol. Chem. 272 (51): 32656–62. doi:10.1074/jbc.272.51.32656. PMID 9405483.
- Ponting CP, Aravind L (April 1999). “START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins”. Trends Biochem. Sci. 24 (4): 130–2. doi:10.1016/S0968-0004(99)01362-6. PMID 10322415.
- Alpy F, Tomasetto C (June 2006). “MLN64 and MENTHO, two mediators of endosomal cholesterol transport”. Biochem. Soc. Trans. 34 (Pt 3): 343–5. doi:10.1042/BST0340343. PMID 16709157.
- Stocco DM, Wang X, Jo Y, Manna PR (November 2005). “Multiple Signaling Pathways Regulating Steroidogenesis and Steroidogenic Acute Regulatory Protein Expression: More Complicated than We Thought”. Molecular Endocrinology. 19 (11): 2647–59. doi:10.1210/me.2004-0532. PMID 15831519.
- Bhangoo A, Anhalt H, Ten S, King SR (March 2006). “Phenotypic variations in lipoid congenital adrenal hyperplasia”. Pediatr Endocrinol Rev. 3 (3): 258–71. PMID 16639391.
- Srivastava VK, Vijayan E, Hiney JK, Dees WL (October 2005). “Effect of ethanol on follicle stimulating hormone-induced steroidogenic acute regulatory protein (StAR) in cultured rat granulosa cells”. Alcohol. 37 (2): 105–11. doi:10.1016/j.alcohol.2006.01.001. PMID 16584974.
- Kariyazono Y, Taura J, Hattori Y, Ishii Y, Narimatsu S, Fujimura M, Takeda T, Yamada H (December 2015). “Effect of in utero exposure to endocrine disruptors on fetal steroidogenesis governed by the pituitary-gonad axis: a study in rats using different ways of administration”. The Journal of Toxicological Sciences. 40 (6): 909–16. doi:10.2131/jts.40.909. PMID 26558472.
- Motohashi M, Wempe MF, Mutou T, Okayama Y, Kansaku N, Takahashi H, Ikegami M, Asari M, Wakui S (2016). “In utero-exposed di(n-butyl) phthalate induce dose dependent, age-related changes of morphology and testosterone-biosynthesis enzymes/associated proteins of Leydig cell mitochondria in rats”. The Journal of Toxicological Sciences. 41 (2): 195–206. doi:10.2131/jts.41.195. PMID 26961603.
- Jin Y, Liu J, Wang L, Chen R, Zhou C, Yang Y, Liu W, Fu Z (2012). “Permethrin exposure during puberty has the potential to enantioselectively induce reproductive toxicity in mice”. Environment International. 42: 144–151. doi:10.1016/j.envint.2011.05.020. ISSN 0160-4120. PMID 21745691.
- Wang H, Wang Q, Zhao X, Liu P, Meng X, Yu T, Ji Y, Zhang H, Zhang C, Zhang Y, Xu D (2009). “Cypermethrin exposure during puberty disrupts testosterone synthesis via downregulating StAR in mouse testes”. Archives of Toxicology. 84 (1): 53–61. doi:10.1007/s00204-009-0479-y. ISSN 0340-5761. PMID 19862501. S2CID 22210562.
- Clark BJ, Cochrum RK (2007). “The Steroidogenic Acute Regulatory Protein as a Target of Endocrine Disruption in Male Reproduction”. Drug Metabolism Reviews. 39 (2–3): 353–370. doi:10.1080/03602530701519151. PMID 17786626. S2CID 26531354.
- Baker BY, Lin L, Kim CJ, Raza J, Smith CP, Miller WL, Achermann JC (December 2006). “Nonclassic congenital lipoid adrenal hyperplasia: a new disorder of the steroidogenic acute regulatory protein with very late presentation and normal male genitalia”. J. Clin. Endocrinol. Metab. 91 (12): 4781–5. doi:10.1210/jc.2006-1565. PMC 1865081. PMID 16968793.
- Metherell LA, Naville D, Halaby G, Begeot M, Huebner A, Nürnberg G, Nürnberg P, Green J, Tomlinson JW, Krone NP, Lin L, Racine M, Berney DM, Achermann JC, Arlt W, Clark AJ (October 2009). “Nonclassic lipoid congenital adrenal hyperplasia masquerading as familial glucocorticoid deficiency”. J. Clin. Endocrinol. Metab. 94 (10): 3865–3871. doi:10.1210/jc.2009-0467. PMC 2860769. PMID 19773404.
- Hutson JC (January 2006). “Physiologic interactions between macrophages and Leydig cells”. Exp. Biol. Med. (Maywood). 231 (1): 1–7. doi:10.1177/153537020623100101. PMID 16380639. S2CID 43006988.
- Hall EA, Ren S, Hylemon PB, Rodriguez-Agudo D, Redford K, Marques D, Kang D, Gil G, Pandak WM (April 2005). “Detection of the steroidogenic acute regulatory protein, StAR, in human liver cells”. Biochim Biophys Acta. 1733 (2–3): 111–119. doi:10.1016/j.bbalip.2005.01.004. PMID 15863358.
- Ma Y, Ren S, Pandak WM, Li X, Ning Y, Lu C, Zhao F, Yin L (December 2007). “The effects of inflammatory cytokines on steroidogenic acute regulatory protein expression in macrophages”. Inflamm Res. 56 (12): 495–501. doi:10.1007/s00011-007-6133-3. PMID 18210233. S2CID 21308251.
- Taylor JM, Borthwick F, Bartholomew C, Graham A (June 2010). “Overexpression of steroidogenic acute regulatory protein increases macrophage cholesterol efflux to apolipoprotein AI”. Cardiovasc Res. 86 (3): 526–534. doi:10.1093/cvr/cvq015. PMID 20083572.
- Anuka E, Yivgi-Ohana N, Eimerl S, Garfinkel B, Melamed-Book N, Chepurkol E, Aravot D, Zinman T, Shainberg A, Hochhauser E, Orly J (September 2013). “Infarct-Induced Steroidogenic Acute Regulatory Protein: A Survival Role in Cardiac Fibroblasts”. Mol Endocrinol. 27 (9): 1502–1517. doi:10.1210/me.2013-1006. PMC 5415234. PMID 23831818.
- Clark BJ, Wells J, King SR, Stocco DM (November 1994). “The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR)”. J Biol Chem. 269 (45): 28314–28322. doi:10.1016/S0021-9258(18)46930-X. PMID 7961770.
- Lin D, Sugawara T, Strauss III JF, Clark BJ, Stocco DM, Saenger P, Rogol A, Miller WL (March 1995). “Role of steroidogenic acute regulatory protein in adrenal and gonadal steroidogenesis”. Science. 267 (5205): 1828–1831. Bibcode:1995Sci…267.1828L. doi:10.1126/science.7892608. PMID 7892608.
- Krueger RJ, Orme-Johnson NR (August 1983). “Acute adrenocorticotropic hormone stimulation of adrenal corticosteroidogenesis”. J Biol Chem. 258 (16): 10159–10167. doi:10.1016/S0021-9258(17)44619-9. PMID 6309771.
- GRCh38: Ensembl release 89: ENSG00000131748 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000018167 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Entrez Gene: StAR related lipid transfer domain containing 3”. Retrieved 2018-08-07.
- Alpy F, Tomasetto C (June 2006). “MLN64 and MENTHO, two mediators of endosomal cholesterol transport”. Biochemical Society Transactions. 34 (Pt 3): 343–5. doi:10.1042/BST0340343. PMID 16709157.
- Wilhelm LP, Wendling C, Védie B, Kobayashi T, Chenard MP, Tomasetto C, Drin G, Alpy F (May 2017). “STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites”. The EMBO Journal. 36 (10): 1412–1433. doi:10.15252/embj.201695917. PMC 5430228. PMID 28377464.
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio MC, Levine TP, Tomasetto C (December 2013). “STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER”. Journal of Cell Science. 126 (Pt 23): 5500–12. doi:10.1242/jcs.139295. PMID 24105263.
- Wilhelm LP, Wendling C, Védie B, Kobayashi T, Chenard MP, Tomasetto C, Drin G, Alpy F (May 2017). “STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites”. The EMBO Journal. 36 (10): 1412–1433. doi:10.15252/embj.201695917. PMC 5430228. PMID 28377464.
- Alpy F, Stoeckel ME, Dierich A, Escola JM, Wendling C, Chenard MP, Vanier MT, Gruenberg J, Tomasetto C, Rio MC (February 2001). “The steroidogenic acute regulatory protein homolog MLN64, a late endosomal cholesterol-binding protein”. The Journal of Biological Chemistry. 276 (6): 4261–9. doi:10.1074/jbc.M006279200. PMID 11053434.
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio MC, Levine TP, Tomasetto C (December 1, 2013). “STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER” (PDF). J Cell Sci. 126 (23): 5500–5512. doi:10.1242/jcs.139295. PMID 24105263. S2CID 7245863.
- Di Mattia, Thomas; Martinet, Arthur; Ikhlef, Souade; McEwen, Alastair G; Nominé, Yves; Wendling, Corinne; Poussin-Courmontagne, Pierre; Voilquin, Laetitia; Eberling, Pascal; Ruffenach, Frank; Cavarelli, Jean; Slee, John; Levine, Timothy P; Drin, Guillaume; Tomasetto, Catherine; Alpy, Fabien (December 1, 2020). “FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts”. The EMBO Journal. 39 (23): e104369. doi:10.15252/embj.2019104369. ISSN 0261-4189. PMC 7705450. PMID 33124732.
- Watari H, Arakane F, Moog-Lutz C, Kallen CB, Tomasetto C, Gerton GL, Rio MC, Baker ME, Strauss JF (August 1997). “MLN64 contains a domain with homology to the steroidogenic acute regulatory protein (StAR) that stimulates steroidogenesis”. Proceedings of the National Academy of Sciences of the United States of America. 94 (16): 8462–7. Bibcode:1997PNAS…94.8462W. doi:10.1073/pnas.94.16.8462. PMC 22957. PMID 9237999.
- Li B, Vachali P, Frederick JM, Bernstein PS (April 2011). “Identification of StARD3 as a lutein-binding protein in the macula of the primate retina”. Biochemistry. 50 (13): 2541–9. doi:10.1021/bi101906y. PMC 3070171. PMID 21322544.
- Alpy F, Wendling C, Rio MC, Tomasetto C (December 2002). “MENTHO, a MLN64 homologue devoid of the START domain”. The Journal of Biological Chemistry. 277 (52): 50780–7. doi:10.1074/jbc.M208290200. PMID 12393907.
- Alpy F, Latchumanan VK, Kedinger V, Janoshazi A, Thiele C, Wendling C, Rio MC, Tomasetto C (May 2005). “Functional characterization of the MENTAL domain”. The Journal of Biological Chemistry. 280 (18): 17945–52. doi:10.1074/jbc.M500723200. PMID 15718238.
- Tsujishita Y, Hurley JH (May 2000). “Structure and lipid transport mechanism of a StAR-related domain”. Nature Structural Biology. 7 (5): 408–14. doi:10.1038/75192. PMID 10802740. S2CID 10806665.
- King SR, Smith AG, Alpy F, Tomasetto C, Ginsberg SD, Lamb DJ (2006). “Characterization of the putative cholesterol transport protein metastatic lymph node 64 in the brain”. Neuroscience. 139 (3): 1031–8. doi:10.1016/j.neuroscience.2006.01.063. PMID 16549269. S2CID 33113555.
- Kishida T, Kostetskii I, Zhang Z, Martinez F, Liu P, Walkley SU, Dwyer NK, Blanchette-Mackie EJ, Radice GL, Strauss JF (April 2004). “Targeted mutation of the MLN64 START domain causes only modest alterations in cellular sterol metabolism”. The Journal of Biological Chemistry. 279 (18): 19276–85. doi:10.1074/jbc.M400717200. PMID 14963026.
- Zhang M, Liu P, Dwyer NK, Christenson LK, Fujimoto T, Martinez F, Comly M, Hanover JA, Blanchette‐Mackie EJ, Strauss JF (2002) MLN64 mediates mobilization of lysosomal cholesterol to steroidogenic mitochondria. J Biol Chem 277: 33300–33310 [PubMed] doi: 10.1074/jbc.M200003200
- ENSG00000288362 GRCh38: Ensembl release 89: ENSG00000140459, ENSG00000288362 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000032323 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- Hanukoglu I (December 1992). “Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis”. The Journal of Steroid Biochemistry and Molecular Biology. 43 (8): 779–804. doi:10.1016/0960-0760(92)90307-5. PMID 22217824. S2CID 112729.
- “Entrez Gene: CYP11A1 cytochrome P450, family 11, subfamily A, polypeptide 1”.
- Strauss JF, Martinez F, Kiriakidou M (February 1996). “Placental steroid hormone synthesis: unique features and unanswered questions”. Biology of Reproduction. 54 (2): 303–311. doi:10.1095/biolreprod54.2.303. PMID 8788180.
- Stoffel-Wagner B (December 2001). “Neurosteroid metabolism in the human brain”. European Journal of Endocrinology. 145 (6): 669–679. doi:10.1530/eje.0.1450669. PMID 11720889.
- Hanukoglu I, Hanukoglu Z (May 1986). “Stoichiometry of mitochondrial cytochromes P-450, adrenodoxin and adrenodoxin reductase in adrenal cortex and corpus luteum. Implications for membrane organization and gene regulation”. European Journal of Biochemistry. 157 (1): 27–31. doi:10.1111/j.1432-1033.1986.tb09633.x. PMID 3011431.
- Hanukoglu I, Suh BS, Himmelhoch S, Amsterdam A (October 1990). “Induction and mitochondrial localization of cytochrome P450scc system enzymes in normal and transformed ovarian granulosa cells”. The Journal of Cell Biology. 111 (4): 1373–1381. doi:10.1083/jcb.111.4.1373. PMC 2116250. PMID 2170421.
- Hanukoglu I, Feuchtwanger R, Hanukoglu A (November 1990). “Mechanism of corticotropin and cAMP induction of mitochondrial cytochrome P450 system enzymes in adrenal cortex cells”. The Journal of Biological Chemistry. 265 (33): 20602–20608. doi:10.1016/S0021-9258(17)30545-8. PMID 2173715.
- Topological studies of cytochromes P-450scc and P-45011 beta in bovine adrenocortical inner mitochondrial membranes. Effects of controlled tryptic digestion. J. Biol. Chem. 1979 254: 10443-8.
- Farkash Y, Timberg R, Orly J (April 1986). “Preparation of antiserum to rat cytochrome P-450 cholesterol side chain cleavage, and its use for ultrastructural localization of the immunoreactive enzyme by protein A-gold technique”. Endocrinology. 118 (4): 1353–1365. doi:10.1210/endo-118-4-1353. PMID 3948785.
- Hanukoglu I, Privalle CT, Jefcoate CR (May 1981). “Mechanisms of ionic activation of adrenal mitochondrial cytochromes P-450scc and P-45011 beta”. The Journal of Biological Chemistry. 256 (9): 4329–4335. doi:10.1016/S0021-9258(19)69437-8. PMID 6783659.
- Hanukoglu I, Rapoport R (1995). “Routes and regulation of NADPH production in steroidogenic mitochondria”. Endocrine Research. 21 (1–2): 231–241. doi:10.3109/07435809509030439. PMID 7588385.
- Hanukoglu I, Gutfinger T, Haniu M, Shively JE (December 1987). “Isolation of a cDNA for adrenodoxin reductase (ferredoxin-NADP+ reductase). Implications for mitochondrial cytochrome P-450 systems”. European Journal of Biochemistry. 169 (3): 449–455. doi:10.1111/j.1432-1033.1987.tb13632.x. PMID 3691502.
- Hanukoglu I, Jefcoate CR (April 1980). “Mitochondrial cytochrome P-450scc. Mechanism of electron transport by adrenodoxin”. The Journal of Biological Chemistry. 255 (7): 3057–3061. doi:10.1016/S0021-9258(19)85851-9. PMID 6766943.
- Hanukoglu I, Spitsberg V, Bumpus JA, Dus KM, Jefcoate CR (May 1981). “Adrenal mitochondrial cytochrome P-450scc. Cholesterol and adrenodoxin interactions at equilibrium and during turnover”. The Journal of Biological Chemistry. 256 (9): 4321–4328. doi:10.1016/S0021-9258(19)69436-6. PMID 7217084.
- Strushkevich N, MacKenzie F, Cherkesova T, Grabovec I, Usanov S, Park HW (June 2011). “Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system”. Proceedings of the National Academy of Sciences of the United States of America. 108 (25): 10139–10143. Bibcode:2011PNAS..10810139S. doi:10.1073/pnas.1019441108. PMC 3121847. PMID 21636783.
- Hanukoglu I, Rapoport R, Weiner L, Sklan D (September 1993). “Electron leakage from the mitochondrial NADPH-adrenodoxin reductase-adrenodoxin-P450scc (cholesterol side chain cleavage) system”. Archives of Biochemistry and Biophysics. 305 (2): 489–498. doi:10.1006/abbi.1993.1452. PMID 8396893.
- Hanukoglu I (2006). “Antioxidant protective mechanisms against reactive oxygen species (ROS) generated by mitochondrial P450 systems in steroidogenic cells”. Drug Metabolism Reviews. 38 (1–2): 171–196. doi:10.1080/03602530600570040. PMID 16684656. S2CID 10766948.
- Lavoie HA, King SR (August 2009). “Transcriptional regulation of steroidogenic genes: STARD1, CYP11A1 and HSD3B”. Experimental Biology and Medicine. 234 (8): 880–907. doi:10.3181/0903-MR-97. PMID 19491374. S2CID 5350278.
- Guo IC, Shih MC, Lan HC, Hsu NC, Hu MC, Chung BC (July 2007). “Transcriptional regulation of human CYP11A1 in gonads and adrenals”. Journal of Biomedical Science. 14 (4): 509–515. doi:10.1007/s11373-007-9177-z. PMID 17594537.
- Hanukoglu A, Fried D, Nakash I, Hanukoglu I (November 1995). “Selective increases in adrenal steroidogenic capacity during acute respiratory disease in infants”. European Journal of Endocrinology. 133 (5): 552–556. doi:10.1530/eje.0.1330552. PMID 7581984. S2CID 44439040.
- Bhangoo A, Anhalt H, Ten S, King SR (March 2006). “Phenotypic variations in lipoid congenital adrenal hyperplasia”. Pediatric Endocrinology Reviews. 3 (3): 258–271. PMID 16639391.
- al Kandari H, Katsumata N, Alexander S, Rasoul MA (August 2006). “Homozygous mutation of P450 side-chain cleavage enzyme gene (CYP11A1) in 46, XY patient with adrenal insufficiency, complete sex reversal, and agenesis of corpus callosum”. The Journal of Clinical Endocrinology and Metabolism. 91 (8): 2821–2826. doi:10.1210/jc.2005-2230. PMID 16705068.
- Kim CJ, Lin L, Huang N, Quigley CA, AvRuskin TW, Achermann JC, Miller WL (March 2008). “Severe combined adrenal and gonadal deficiency caused by novel mutations in the cholesterol side chain cleavage enzyme, P450scc”. The Journal of Clinical Endocrinology and Metabolism. 93 (3): 696–702. doi:10.1210/jc.2007-2330. PMC 2266942. PMID 18182448.
- FBuonocore F, Achermann JC (2020). “Primary adrenal insufficiency: New genetic causes and their long-term consequences”. Clinical Endocrinology. 92 (1): 11–20. doi:10.1111/cen.14109. PMC 6916405. PMID 31610036.
- Becker KL (2001). Principles and Practice of Endocrinology and Metabolism. Lippincott Williams & Wilkins. pp. 735–. ISBN 978-0-7817-1750-2.
- Jameson JL, De Groot LJ (18 May 2010). Endocrinology – E-Book: Adult and Pediatric. Elsevier Health Sciences. pp. 301–302. ISBN 978-1-4557-1126-0.
- Ortiz de Montellano PR (13 March 2015). Cytochrome P450: Structure, Mechanism, and Biochemistry. Springer. pp. 851–879. ISBN 978-3-319-12108-6.
Further reading
- Morohashi KI, Omura T (December 1996). “Ad4BP/SF-1, a transcription factor essential for the transcription of steroidogenic cytochrome P450 genes and for the establishment of the reproductive function”. FASEB Journal. 10 (14): 1569–77. doi:10.1096/fasebj.10.14.9002548. PMID 9002548. S2CID 13891159.
- Achermann JC, Meeks JJ, Jameson JL (December 2001). “Phenotypic spectrum of mutations in DAX-1 and SF-1”. Molecular and Cellular Endocrinology. 185 (1–2): 17–25. doi:10.1016/S0303-7207(01)00619-0. PMID 11738790. S2CID 20651430.
- Ozisik G, Achermann JC, Jameson JL (June 2002). “The role of SF1 in adrenal and reproductive function: insight from naturally occurring mutations in humans”. Molecular Genetics and Metabolism. 76 (2): 85–91. doi:10.1016/S1096-7192(02)00032-X. PMID 12083805.
- de-Souza BF, Lin L, Achermann JC (June 2006). “Steroidogenic factor-1 (SF-1) and its relevance to pediatric endocrinology”. Pediatric Endocrinology Reviews. 3 (4): 359–64. doi:10.1159/000094108. PMID 16816804.
- Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA, Tourtellotte LM, Simburger K, Milbrandt J (November 1995). “Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids”. Proceedings of the National Academy of Sciences of the United States of America. 92 (24): 10939–43. Bibcode:1995PNAS…9210939S. doi:10.1073/pnas.92.24.10939. PMC 40546. PMID 7479914.
- Sasano H, Shizawa S, Suzuki T, Takayama K, Fukaya T, Morohashi K, Nagura H (August 1995). “Ad4BP in the human adrenal cortex and its disorders”. The Journal of Clinical Endocrinology and Metabolism. 80 (8): 2378–80. doi:10.1210/jcem.80.8.7629233. PMID 7629233.
- Oba K, Yanase T, Nomura M, Morohashi K, Takayanagi R, Nawata H (September 1996). “Structural characterization of human Ad4bp (SF-1) gene”. Biochemical and Biophysical Research Communications. 226 (1): 261–7. doi:10.1006/bbrc.1996.1343. PMID 8806624.
- Asa SL, Bamberger AM, Cao B, Wong M, Parker KL, Ezzat S (June 1996). “The transcription activator steroidogenic factor-1 is preferentially expressed in the human pituitary gonadotroph”. The Journal of Clinical Endocrinology and Metabolism. 81 (6): 2165–70. doi:10.1210/jcem.81.6.8964846. PMID 8964846.
- Bamberger AM, Ezzat S, Cao B, Wong M, Parker KL, Schulte HM, Asa SL (June 1996). “Expression of steroidogenic factor-1 (SF-1) mRNA and protein in the human placenta”. Molecular Human Reproduction. 2 (6): 457–61. doi:10.1093/molehr/2.6.457. PMID 9238716.
- Crawford PA, Polish JA, Ganpule G, Sadovsky Y (October 1997). “The activation function-2 hexamer of steroidogenic factor-1 is required, but not sufficient for potentiation by SRC-1”. Molecular Endocrinology. 11 (11): 1626–35. doi:10.1210/mend.11.11.9970. PMID 9328345. S2CID 35590139.
- Nachtigal MW, Hirokawa Y, Enyeart-VanHouten DL, Flanagan JN, Hammer GD, Ingraham HA (May 1998). “Wilms’ tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression”. Cell. 93 (3): 445–54. doi:10.1016/S0092-8674(00)81172-1. PMID 9590178. S2CID 19015882.
- Hammer GD, Krylova I, Zhang Y, Darimont BD, Simpson K, Weigel NL, Ingraham HA (April 1999). “Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress”. Molecular Cell. 3 (4): 521–6. doi:10.1016/S1097-2765(00)80480-3. PMID 10230405.
- Achermann JC, Ito M, Ito M, Hindmarsh PC, Jameson JL (June 1999). “A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans”. Nature Genetics. 22 (2): 125–6. doi:10.1038/9629. PMID 10369247. S2CID 27674149.
- Dugger BN, Morris JA, Jordan CL, Breedlove SM (February 2007). “Androgen receptors are required for full masculinization of the ventromedial hypothalamus (VMH) in rats”. Hormones and Behavior. 51 (2): 195–201. doi:10.1016/j.yhbeh.2006.10.001. PMC 1828277. PMID 17123532.
- Storlien LH, Albert DJ (August 1972). “The effect of VMH lesions, lateral cuts and anterior cuts of food intake, activity level, food motivation, and reactivity to taste”. Physiology & Behavior. 9 (2): 191–7. doi:10.1016/0031-9384(72)90234-x. PMID 4569944.
- Carlson N (2010). Physiology of behavior (10th ed.). Boston: Allyn and Bacon. pp. 355–357.
- Zhang M, Liu P, Dwyer NK, Christenson LK, Fujimoto T, Martinez F, Comly M, Hanover JA, Blanchette-Mackie EJ, Strauss JF (September 2002). “MLN64 mediates mobilization of lysosomal cholesterol to steroidogenic mitochondria”. The Journal of Biological Chemistry. 277 (36): 33300–10. doi:10.1074/jbc.M200003200. PMID 12070139.
- Strauss JF, Liu P, Christenson LK, Watari H (November 2002). “Sterols and intracellular vesicular trafficking: lessons from the study of NPC1”. Steroids. 67 (12): 947–51. doi:10.1016/s0039-128x(02)00042-9. PMID 12398991. S2CID 25185703.
- Tuckey RC, Bose HS, Czerwionka I, Miller WL (April 2004). “Molten globule structure and steroidogenic activity of N-218 MLN64 in human placental mitochondria”. Endocrinology. 145 (4): 1700–7. doi:10.1210/en.2003-1034. PMID 14715710.
- Katoh M, Katoh M (April 2004). “Evolutionary recombination hotspot around GSDML-GSDM locus is closely linked to the oncogenomic recombination hotspot around the PPP1R1B-ERBB2-GRB7 amplicon”. International Journal of Oncology. 24 (4): 757–63. doi:10.3892/ijo.24.4.757. PMID 15010812.
- Alpy F, Latchumanan VK, Kedinger V, Janoshazi A, Thiele C, Wendling C, Rio MC, Tomasetto C (May 2005). “Functional characterization of the MENTAL domain”. The Journal of Biological Chemistry. 280 (18): 17945–52. doi:10.1074/jbc.M500723200. PMID 15718238.
- Hölttä-Vuori M, Alpy F, Tanhuanpää K, Jokitalo E, Mutka AL, Ikonen E (August 2005). “MLN64 is involved in actin-mediated dynamics of late endocytic organelles”. Molecular Biology of the Cell. 16 (8): 3873–86. doi:10.1091/mbc.e04-12-1105. PMC 1182323. PMID 15930133.
- Alpy F, Tomasetto C (June 2006). “MLN64 and MENTHO, two mediators of endosomal cholesterol transport”. Biochemical Society Transactions. 34 (Pt 3): 343–5. doi:10.1042/BST0340343. PMID 16709157.
- Murcia M, Faráldo-Gómez JD, Maxfield FR, Roux B (December 2006). “Modeling the structure of the StART domains of MLN64 and StAR proteins in complex with cholesterol”. Journal of Lipid Research. 47 (12): 2614–30. doi:10.1194/jlr.M600232-JLR200. PMID 16990645.
- Benusiglio PR, Pharoah PD, Smith PL, Lesueur F, Conroy D, Luben RN, Dew G, Jordan C, Dunning A, Easton DF, Ponder BA (December 2006). “HapMap-based study of the 17q21 ERBB2 amplicon in susceptibility to breast cancer”. British Journal of Cancer. 95 (12): 1689–95. doi:10.1038/sj.bjc.6603473. PMC 2360759. PMID 17117180.
- Helmberg A (August 1993). “Twin genes and endocrine disease: CYP21 and CYP11B genes”. Acta Endocrinologica. 129 (2): 97–108. doi:10.1530/acta.0.1290097. PMID 8372604.
- Papadopoulos V, Amri H, Boujrad N, Cascio C, Culty M, Garnier M, et al. (January 1997). “Peripheral benzodiazepine receptor in cholesterol transport and steroidogenesis”. Steroids. 62 (1): 21–28. doi:10.1016/S0039-128X(96)00154-7. PMID 9029710. S2CID 1977513.
- Stocco DM (June 2000). “Intramitochondrial cholesterol transfer”. Biochimica et Biophysica Acta (BBA) – Molecular and Cell Biology of Lipids. 1486 (1): 184–197. doi:10.1016/S1388-1981(00)00056-1. PMID 10856721.
- Kristensen VN, Kure EH, Erikstein B, Harada N, Børresen-Dale A (October 2001). “Genetic susceptibility and environmental estrogen-like compounds”. Mutation Research. 482 (1–2): 77–82. doi:10.1016/S0027-5107(01)00212-3. PMID 11535251.
- Strauss JF (November 2003). “Some new thoughts on the pathophysiology and genetics of polycystic ovary syndrome”. Annals of the New York Academy of Sciences. 997 (1): 42–48. Bibcode:2003NYASA.997…42S. doi:10.1196/annals.1290.005. PMID 14644808. S2CID 23559461.
- Wada A, Waterman MR (November 1992). “Identification by site-directed mutagenesis of two lysine residues in cholesterol side chain cleavage cytochrome P450 that are essential for adrenodoxin binding”. The Journal of Biological Chemistry. 267 (32): 22877–22882. doi:10.1016/S0021-9258(18)50028-4. PMID 1429635.
- Hu MC, Guo IC, Lin JH, Chung BC (March 1991). “Regulated expression of cytochrome P-450scc (cholesterol-side-chain cleavage enzyme) in cultured cell lines detected by antibody against bacterially expressed human protein”. The Biochemical Journal. 274 (Pt 3): 813–817. doi:10.1042/bj2740813. PMC 1149983. PMID 1849407.
- Sparkes RS, Klisak I, Miller WL (June 1991). “Regional mapping of genes encoding human steroidogenic enzymes: P450scc to 15q23-q24, adrenodoxin to 11q22; adrenodoxin reductase to 17q24-q25; and P450c17 to 10q24-q25”. DNA and Cell Biology. 10 (5): 359–365. doi:10.1089/dna.1991.10.359. PMID 1863359.
- Coghlan VM, Vickery LE (October 1991). “Site-specific mutations in human ferredoxin that affect binding to ferredoxin reductase and cytochrome P450scc”. The Journal of Biological Chemistry. 266 (28): 18606–18612. doi:10.1016/S0021-9258(18)55106-1. PMID 1917982.
- Matteson KJ, Chung BC, Urdea MS, Miller WL (April 1986). “Study of cholesterol side-chain cleavage (20,22 desmolase) deficiency causing congenital lipoid adrenal hyperplasia using bovine-sequence P450scc oligodeoxyribonucleotide probes”. Endocrinology. 118 (4): 1296–1305. doi:10.1210/endo-118-4-1296. PMID 2419119.
- Chung BC, Matteson KJ, Voutilainen R, Mohandas TK, Miller WL (December 1986). “Human cholesterol side-chain cleavage enzyme, P450scc: cDNA cloning, assignment of the gene to chromosome 15, and expression in the placenta”. Proceedings of the National Academy of Sciences of the United States of America. 83 (23): 8962–8966. Bibcode:1986PNAS…83.8962C. doi:10.1073/pnas.83.23.8962. PMC 387054. PMID 3024157.
- Morohashi K, Sogawa K, Omura T, Fujii-Kuriyama Y (April 1987). “Gene structure of human cytochrome P-450(SCC), cholesterol desmolase”. Journal of Biochemistry. 101 (4): 879–887. doi:10.1093/oxfordjournals.jbchem.a121955. PMID 3038854.
- Maruyama K, Sugano S (January 1994). “Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides”. Gene. 138 (1–2): 171–174. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Gharani N, Waterworth DM, Batty S, White D, Gilling-Smith C, Conway GS, et al. (March 1997). “Association of the steroid synthesis gene CYP11a with polycystic ovary syndrome and hyperandrogenism”. Human Molecular Genetics. 6 (3): 397–402. doi:10.1093/hmg/6.3.397. PMID 9147642.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). “Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library”. Gene. 200 (1–2): 149–156. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Hukkanen J, Mäntylä M, Kangas L, Wirta P, Hakkola J, Paakki P, et al. (February 1998). “Expression of cytochrome P450 genes encoding enzymes active in the metabolism of tamoxifen in human uterine endometrium”. Pharmacology & Toxicology. 82 (2): 93–97. doi:10.1111/j.1600-0773.1998.tb01404.x. PMID 9498238.
- Zhou Z, Shackleton CH, Pahwa S, White PC, Speiser PW (March 1998). “Prominent sex steroid metabolism in human lymphocytes”. Molecular and Cellular Endocrinology. 138 (1–2): 61–69. doi:10.1016/S0303-7207(98)00052-5. PMID 9685215. S2CID 20490901.
External links
- GeneReviews/NCBI/NIH/UW entry on 46,XY Disorder of Sex Development and 46,XY Complete Gonadal Dysgenesis
- OMIM entries on 46,XY Disorder of Sex Development and 46,XY Complete Gonadal Dysgenesis
- steroidogenic+factor+1 at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Diagram of Ventromedial motor tract at lemoyne.edu
- New Scientist: Seat of female libido revealed (June 26, 2006)
- steroidogenic+acute+regulatory+protein at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Cytochrome+P450scc at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
| Transcription factors and intracellular receptors | |
|---|---|
| Basic domains | |
| Zinc finger DNA-binding domains | |
| Helix-turn-helix domains | |
| β-Scaffold factors with minor groove contacts | |
| Other transcription factors | |
| see also transcription factor/coregulator deficiencies |
| Metabolism: lipid metabolism – ketones/cholesterol synthesis enzymes/steroid metabolism |
|---|
| Cytochromes, oxygenases: cytochrome P450 (Most belong to EC 1.14) |
|---|
| Mitochondrial proteins |
|---|
| Oxidoreductases: CH-NH (EC 1.5) |
|---|


Leave a Reply