Not to be confused with glycopeptide, proteoglycan, or glycoprotein
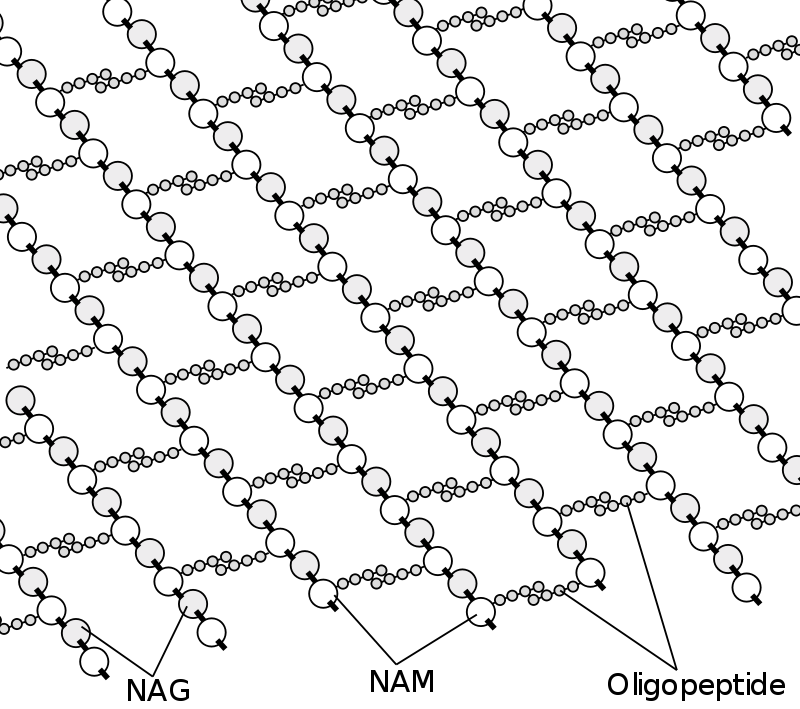
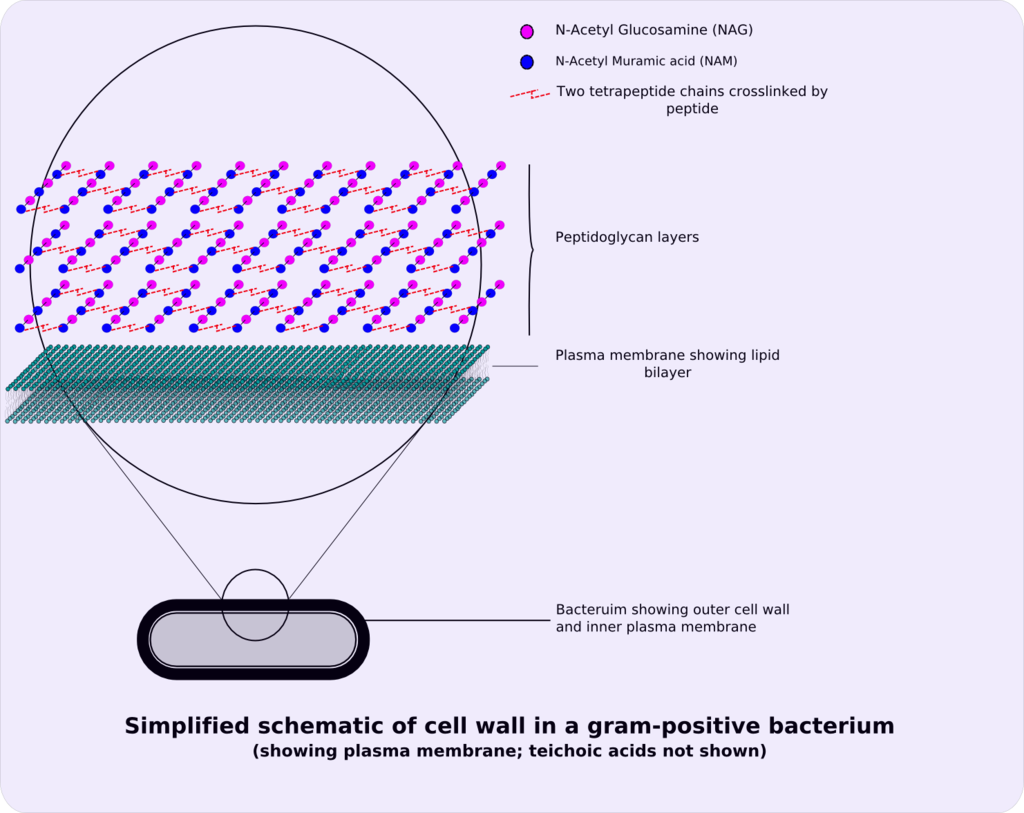
Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like peptidoglycan layer outside the plasma membrane, the rigid cell wall (murein sacculus) characteristic of most bacteria (domain Bacteria).
- Woese CR, Kandler O, Wheelis ML (June 1990). “Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya”. Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–4579. Bibcode:1990PNAS…87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- Madigan, Michael T.; Martinko, John M.; Bender, Kelly S.; Buckley, Daniel H.; Stahl, David A. (2015). Brock Biology of Microorganisms (14 ed.). Boston: Pearson Education Limited. pp. 66–67. ISBN 978-1-292-01831-7.]: 66–67
The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer.
- Madigan, Michael T.; Martinko, John M.; Bender, Kelly S.; Buckley, Daniel H.; Stahl, David A. (2015). Brock Biology of Microorganisms (14 ed.). Boston: Pearson Education Limited. pp. 66–67. ISBN 978-1-292-01831-7.
- Mehta A (20 March 2011). “Animation of Synthesis of Peptidoglycan Layer”. PharmaXChange.info.
Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.
- Belgrave AM, Wolgemuth CW (June 2013). “Elasticity and biochemistry of growth relate replication rate to cell length and cross-link density in rod-shaped bacteria”. Biophysical Journal. 104 (12): 2607–2611. Bibcode:2013BpJ…104.2607B. doi:10.1016/j.bpj.2013.04.028. PMC 3686348. PMID 23790368.
The peptidoglycan layer is substantially thicker in Gram-positive bacteria (20 to 80 nanometers) than in Gram-negative bacteria (7 to 8 nanometers). Depending on pH growth conditions, the peptidoglycan forms around 40 to 90% of the cell wall‘s dry weight of Gram-positive bacteria but only around 10% of Gram-negative strains. Thus, presence of high levels of peptidoglycan is the primary determinant of the characterisation of bacteria as Gram-positive. In Gram-positive strains, it is important in attachment roles and serotyping purposes. For both Gram-positive and Gram-negative bacteria, particles of approximately 2 nm can pass through the peptidoglycan.
- Purcell A (18 March 2016). “Bacteria”. Basic Biology.
- Hogan CM (12 October 2014). “Bacteria”. In Draggan S, Cleveland CJ (eds.). Encyclopedia of Earth. Washington DC: National Council for Science and the Environment.
- Salton MR, Kim KS (1996). “Structure”. In Baron S, et al. (eds.). Structure. In: Baron’s Medical Microbiology (4th ed.). Univ of Texas Medical Branch. ISBN 978-0-9631172-1-2. PMID 21413343.
- Demchick P, Koch AL (February 1996). “The permeability of the wall fabric of Escherichia coli and Bacillus subtilis”. Journal of Bacteriology. 178 (3): 768–773. doi:10.1128/jb.178.3.768-773.1996. PMC 177723. PMID 8550511.
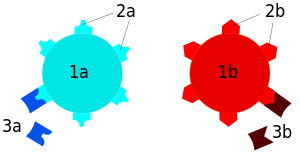
A serotype or serovar is a distinct variation within a species of bacteria or virus or among immune cells of different individuals. These microorganisms, viruses, or cells are classified together based on their surface antigens, allowing the epidemiologic classification of organisms to the subspecies level.[Baron EJ (1996). Baron S; et al. (eds.). Classification. In: Baron’s Medical Microbiology (4th ed.). Univ of Texas Medical Branch. ISBN 978-0-9631172-1-2. (via NCBI Bookshelf).][Ryan KJ, Ray CG, Sherris JC, eds. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 978-0-8385-8529-0.][“Serovar”. The American Heritage Medical Dictionary. Houghton Mifflin Company. 2007.][clarification needed] A group of serovars with common antigens is called a serogroup or sometimes serocomplex.[clarification needed] Serotyping often plays an essential role in determining species and subspecies. The Salmonella genus of bacteria, for example, has been determined to have over 2600 serotypes. Vibrio cholerae, the species of bacteria that causes cholera, has over 200 serotypes, based on cell antigens. Only two of them have been observed to produce the potent enterotoxin that results in cholera: O1 and O139.[citation needed] Serotypes were discovered by the American microbiologist Rebecca Lancefield in 1933.[Lancefield RC (March 1933). “A Serological Differentiation of Human and Other Groups of Hemolytic Streptococci”. The Journal of Experimental Medicine. 57 (4): 571–95. doi:10.1084/jem.57.4.571. PMC 2132252. PMID 19870148.]
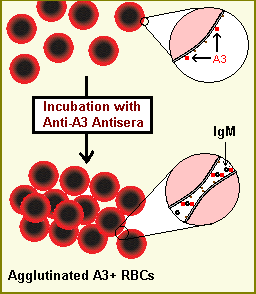
SEROTYPING ROLE IN ORGAN TRANSPLANTATION The immune system is capable of discerning a cell as being ‘self’ or ‘non-self’ according to that cell’s serotype. In humans, that serotype is largely determined by human leukocyte antigen (HLA), the human version of the major histocompatibility complex. Cells determined to be non-self are usually recognized by the immune system as foreign, causing an immune response, such as hemagglutination. Serotypes differ widely between individuals; therefore, if cells from one human (or animal) are introduced into another random human, those cells are often determined to be non-self because they do not match the self-serotype. For this reason, transplants between genetically non-identical humans often induce a problematic immune response in the recipient, leading to transplant rejection. In some situations, this effect can be reduced by serotyping both recipient and potential donors to determine the closest HLA match.[Frohn C, Fricke L, Puchta JC, Kirchner H (February 2001). “The effect of HLA-C matching on acute renal transplant rejection”. Nephrology, Dialysis, Transplantation. 16 (2): 355–60. doi:10.1093/ndt/16.2.355. PMID 11158412.]
Human leukocyte antigens
| HLA Locus | # of Serotypes | Broad Antigens | Split Antigens |
|---|---|---|---|
| A | 25 | 4 | 15 |
| B | 50 | 9 | |
| C* | 12 | 1 | |
| DR | 21 | 4 | |
| DQ | 8 | 2 | |
| DP* |
SEROTYPING OF SALMONELLA The Kauffman–White classification scheme is the basis for naming the manifold serovars of Salmonella. To date, more than 2600 different serotypes have been identified.[Gal-Mor O, Boyle EC, Grassl GA (2014). “Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ”. Frontiers in Microbiology. 5: 391. doi:10.3389/fmicb.2014.00391. PMC 4120697. PMID 25136336.] A Salmonella serotype is determined by the unique combination of reactions of cell surface antigens. The “O” antigen is determined by the outermost portion of the lipopolysaccharide and the “H” antigen is based on the flagellar (protein) antigens.[ “Serotypes and the Importance of Serotyping Salmonella”. CDC. Retrieved 16 October 2014.] There are two species of Salmonella: Salmonella bongori and Salmonella enterica. Salmonella enterica can be subdivided into six subspecies. The process to identify the serovar of the bacterium consists of finding the formula of surface antigens which represent the variations of the bacteria. The traditional method for determining the antigen formula is agglutination reactions on slides. The agglutination between the antigen and the antibody is made with a specific antisera, which reacts with the antigen to produce a mass. The antigen O is tested with a bacterial suspension from an agar plate, whereas the antigen H is tested with a bacterial suspension from a broth culture. The scheme classifies the serovar depending on its antigen formula obtained via the agglutination reactions.[Danan C, Fremy S, Moury F, Bohnert ML, Brisabois A (2009). “Determining the serotype of isolated Salmonella strains in the veterinary sector using the rapid slide agglutination test”. J. Reference. 2: 13–8.] Additional serotyping methods and alternative subtyping methodologies have been reviewed by Wattiau et al.[Wattiau P, Boland C, Bertrand S (November 2011). “Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives”. Applied and Environmental Microbiology. 77 (22): 7877–85. Bibcode:2011ApEnM..77.7877W. doi:10.1128/AEM.05527-11. PMC 3209009. PMID 21856826.] See also: Biovar and Morphovar
It is difficult to tell whether an organism is gram-positive or gram-negative using a microscope; gram staining, created by Hans Christian Gram in 1884, is required. The bacteria are stained with several dyes such as crystal violet, iodine alcohol, and safranin using the gram staining procedure. Gram positive cells are purple after staining, while Gram negative cells are red.
- Coico R (October 2005). “Gram Staining”. In Coico R, Kowalik T, Quarles J, Stevenson B (eds.). Current Protocols in Microbiology. Vol. Appendix 3. Hoboken, NJ, USA: John Wiley & Sons, Inc. pp. mca03cs00. doi:10.1002/9780471729259.mca03cs00. ISBN 978-0-471-72925-9. PMID 18770544. S2CID 32452815.
Structure
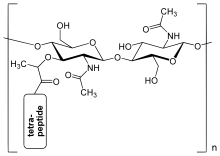
The peptidoglycan layer in the bacterial cell wall is a crystal lattice structure formed from linear chains of two alternating amino sugars, namely N-acetylglucosamine (GlcNAc or NAG) and N-acetylmuramic acid (MurNAc or NAM). The alternating sugars are connected by a β-(1,4)-glycosidic bond. Each MurNAc is attached to a short (4- to 5-residue) amino acid chain, containing L-alanine, D-glutamic acid, meso-diaminopimelic acid, and D-alanine in the case of Escherichia coli (a Gram-negative bacterium) or L-alanine, D-glutamine, L-lysine, and D-alanine with a 5-glycine interbridge between tetrapeptides in the case of Staphylococcus aureus (a Gram-positive bacterium). Peptidoglycan is one of the most important sources of D-amino acids in nature.[citation needed]
By enclosing the inner membrane, the peptidoglycan layer protects the cell from lysis caused by the turgor pressure of the cell. When the cell wall grows, it retains its shape throughout its life, so a rod shape will remain a rod shape, and a spherical shape will remain a spherical shape for life. This happens because the freshly added septal material of synthesis transforms into a hemispherical wall for the offspring cells.
- Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS (December 2008). “Cell shape and cell-wall organization in Gram-negative bacteria”. Proceedings of the National Academy of Sciences of the United States of America. 105 (49): 19282–19287. Bibcode:2008PNAS..10519282H. doi:10.1073/pnas.0805309105. PMC 2592989. PMID 19050072.
Cross-linking between amino acids in different linear amino sugar chains occurs with the help of the enzyme DD-transpeptidase and results in a 3-dimensional structure that is strong and rigid. The specific amino acid sequence and molecular structure vary with the bacterial species.
- Ryan KJ, Ray CG, eds. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 978-0-8385-8529-0.
The different peptidoglycan types of bacterial cell walls and their taxonomic implications have been described. Archaea (domain Archaea) do not contain peptidoglycan (murein). Some Archaea contain pseudopeptidoglycan (pseudomurein, see below).
- Schleifer KH, Kandler O (December 1972). “Peptidoglycan types of bacterial cell walls and their taxonomic implications”. Bacteriological Reviews. 36 (4): 407–477. doi:10.1128/MMBR.36.4.407-477.1972. PMC 408328. PMID 4568761.
- Kandler O, Hippe H (May 1977). “Lack of peptidoglycan in the cell walls of Methanosarcina barkeri”. Archives of Microbiology. 113 (1–2): 57–60. doi:10.1007/BF00428580. PMID 889387. S2CID 19145374.
- Woese CR, Kandler O, Wheelis ML (June 1990). “Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya”. Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–4579. Bibcode:1990PNAS…87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- Kandler O, König H (April 1998). “Cell wall polymers in Archaea (Archaebacteria)”. Cellular and Molecular Life Sciences. 54 (4): 305–308. doi:10.1007/s000180050156. PMID 9614965. S2CID 13527169.
Peptidoglycan is involved in binary fission during bacterial cell reproduction. L-form bacteria and mycoplasmas, both lacking peptidoglycan cell walls, do not proliferate by binary fission, but by a budding mechanism.
- Kandler G, Kandler O (1954). (Article in English available). “[Studies on morphology and multiplication of pleuropneumonia-like organisms and on bacterial L-phase, I. Light microscopy]” [Studies on morphology and multiplication of pleuropneumonia-like organisms and on bacterial L-phase, I. Light microscopy (now mycoplasmas and L-form bacteria)]. Archiv für Mikrobiologie (in German). 21 (2): 178–201. doi:10.1007/BF01816378. PMID 14350641. S2CID 21257985.
- Leaver M, Domínguez-Cuevas P, Coxhead JM, Daniel RA, Errington J (February 2009). [see also Erratum, 23 July 2009, Nature, vol. 460, p.538]. “Life without a wall or division machine in Bacillus subtilis”. Nature. 457 (7231): 849–853. Bibcode:2009Natur.457..849L.
In the course of early evolution, the successive development of boundaries (membranes, walls) protecting first structures of life against their environment must have been essential for the formation of the first cells (cellularisation).
The invention of rigid peptidoglycan (murein) cell walls in bacteria (domain Bacteria[Woese CR, Kandler O, Wheelis ML (June 1990). “Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya”. Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–4579. Bibcode:1990PNAS…87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.]) was probably the prerequisite for their survival, extensive radiation and colonisation of virtually all habitats of the geosphere and hydrosphere.
- Kandler O (1994). “The early diversification of life”. In Bengtson S (ed.). Early Life on Earth. Nobel Symposium 84. New York: Columbia U.P. pp. 221–270. ISBN 9780231080880.
- Kandler O (1998). “The early diversification of life and the origin of the three domains: A proposal”. In Wiegel J, Adams MW (eds.). Thermophiles: The keys to molecular evolution and the origin of life?. London: Taylor and Francis Ltd. pp. 19–31. ISBN 978-0-203-48420-3.
Biosynthesis
The peptidoglycan monomers are synthesized in the cytosol and are then attached to a membrane carrier bactoprenol. Bactoprenol transports peptidoglycan monomers across the cell membrane where they are inserted into the existing peptidoglycan.[“The Prokaryotic Cell: Bacteria”. Archived from the original on 26 July 2010. Retrieved 1 May 2011.]
- In the first step of peptidoglycan synthesis, glutamine, which is an amino acid, donates an amino group to a sugar, fructose 6-phosphate.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] This reaction, catalyzed by EC 2.6.1.16 (GlmS), turns fructose 6-phosphate into glucosamine-6-phosphate.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
- In step two, an acetyl group is transferred from acetyl CoA to the amino group on the glucosamine-6-phosphate creating N-acetyl-glucosamine-6-phosphate.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] This reaction is EC 5.4.2.10, catalyzed by GlmM.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
- In step three of the synthesis process, the N-acetyl-glucosamine-6-phosphate is isomerized, which will change N-acetyl-glucosamine-6-phosphate to N-acetyl-glucosamine-1-phosphate.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] This is EC 2.3.1.157, catalyzed by GlmU.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
- In step 4, the N-acetyl-glucosamine-1-phosphate, which is now a monophosphate, attacks UTP. Uridine triphosphate, which is a pyrimidine nucleotide, has the ability to act as an energy source. In this particular reaction, after the monophosphate has attacked the UTP, an inorganic pyrophosphate is given off and is replaced by the monophosphate, creating UDP-N-acetylglucosamine (2,4). (When UDP is used as an energy source, it gives off an inorganic phosphate.) This initial stage, is used to create the precursor for the NAG in peptidoglycan.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] This is EC 2.7.7.23, also catalyzed by GlmU, which is a bifunctional enzyme.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
- In step 5, some of the UDP-N-acetylglucosamine (UDP-GlcNAc) is converted to UDP-MurNAc (UDP-N-acetylmuramic acid) by the addition of a lactyl group to the glucosamine. Also in this reaction, the C3 hydroxyl group will remove a phosphate from the alpha carbon of phosphoenolpyruvate. This creates what is called an enol derivative.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] EC 2.5.1.7, catalyzed by MurA.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
- In step 6, the enol is reduced to a “lactyl moiety” by NADPH in step six.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] EC 1.3.1.98, catalyzed by MurB.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
- In step 7, the UDP–MurNAc is converted to UDP-MurNAc pentapeptide by the addition of five amino acids, usually including the dipeptide D-alanyl-D-alanine.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] This is a string of three reactions: EC 6.3.2.8 by MurC, EC 6.3.2.9 by MurD, and EC 6.3.2.13 by MurE.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
Each of these reactions requires the energy source ATP.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] This is all referred to as Stage one.
Stage two occurs in the cytoplasmic membrane. It is in the membrane where a lipid carrier called bactoprenol carries peptidoglycan precursors through the cell membrane.
- Undecaprenyl phosphate will attack the UDP-MurNAc penta, creating a PP-MurNac penta, which is now a lipid (lipid I).[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] EC 2.7.8.13 by MraY.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
- UDP-GlcNAc is then transported to MurNAc, creating Lipid-PP-MurNAc penta-GlcNAc (lipid II), a disaccharide, also a precursor to peptidoglycan.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.] EC 2.4.1.227 by MurG.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
- Lipid II is transported across the membrane by flippase (MurJ), a discovery made in 2014 after decades of searching.[Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N (July 2014). “Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis”. Science. 345 (6193): 220–222. Bibcode:2014Sci…345..220S. doi:10.1126/science.1254522. PMC 4163187. PMID 25013077.] Once it is there, it is added to the growing glycan chain by the enzyme peptidoglycan glycosyltransferase (GTase, EC 2.4.1.129). This reaction is known as tranglycosylation. In the reaction, the hydroxyl group of the GlcNAc will attach to the MurNAc in the glycan, which will displace the lipid-PP from the glycan chain.[White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.]
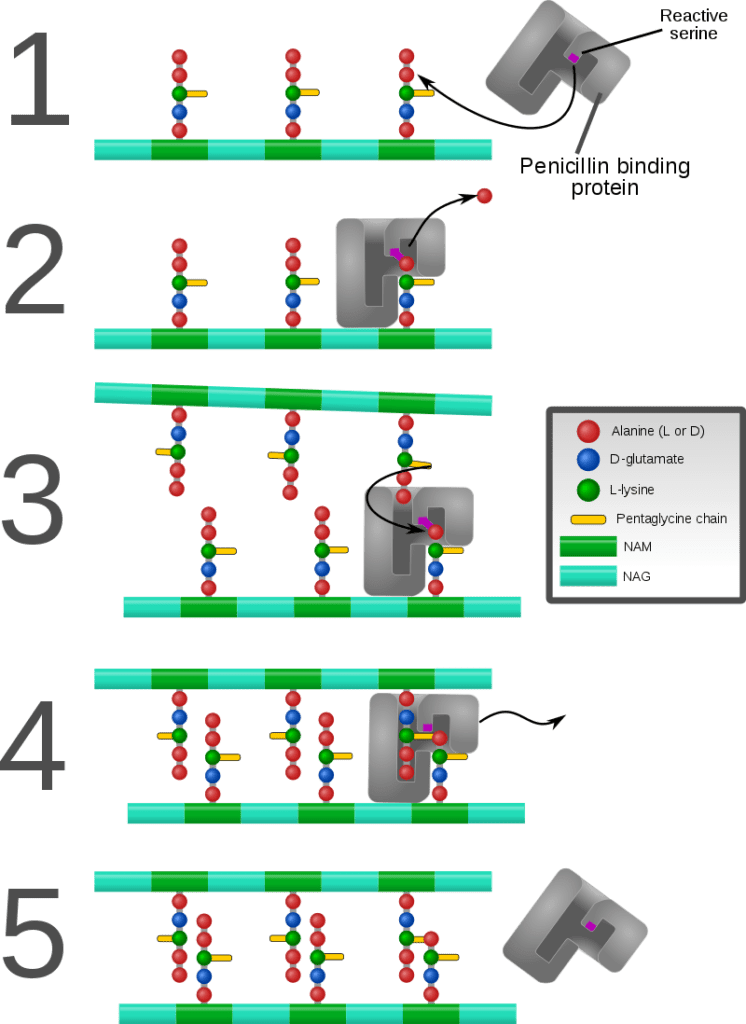
- In a final step, the DD-transpeptidase (TPase, EC 3.4.16.4) crosslinks individual glycan chains. This protein is also known as the penicillin-binding protein. Some versions of the enzyme also performs the glycosyltransferase function, while others leave the job to a separate enzyme.[Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.]
Pseudopeptidoglycan
Main article: Pseudopeptidoglycan
In some archaea, i.e. members of the Methanobacteriales and in the genus Methanopyrus, pseudopeptidoglycan (pseudomurein) has been found. In pseudopeptidoglycan the sugar residues are β-(1,3) linked N-acetylglucosamine and N-acetyltalosaminuronic acid. This makes the cell walls of such archaea insensitive to lysozyme. The biosynthesis of pseudopeptidoglycan has been described.
- Kandler O, König H (April 1998). “Cell wall polymers in Archaea (Archaebacteria)”. Cellular and Molecular Life Sciences. 54 (4): 305–308. doi:10.1007/s000180050156. PMID 9614965. S2CID 13527169.
- Madigan MT, Martinko JM, Dunlap PV, Clark DP (2009). Brock Biology of Microorganisms (12th ed.). San Francisco, CA: Pearson/Benjamin Cummings.
- König H, Kandler O, Hammes W (January 1989). “Biosynthesis of pseudomurein: isolation of putative precursors from Methanobacterium thermoautotrophicum”. Canadian Journal of Microbiology. 35 (1): 176–181. doi:10.1139/m89-027. PMID 2720492.
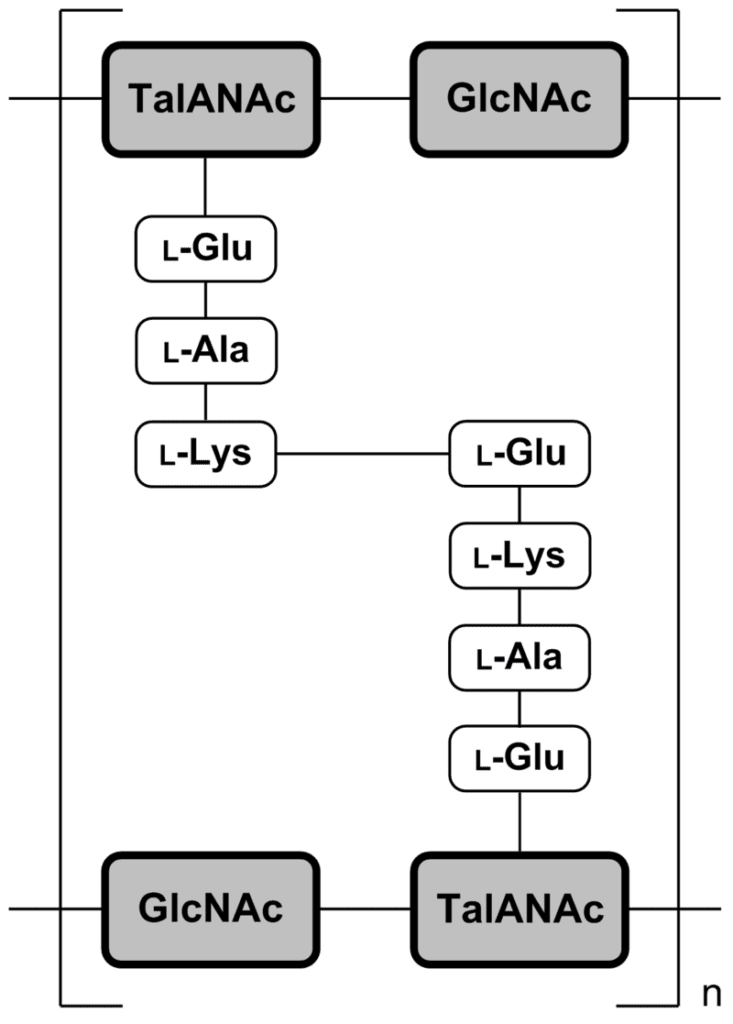
Pseudopeptidoglycan (also known as pseudomurein;[White, David. (1995) The Physiology and Biochemistry of Prokaryotes, pages 6, 12-21. (Oxford: Oxford University Press). ISBN 0-19-508439-X.] PPG hereafter) is a major cell wall component of some Archaea that differs from bacterial peptidoglycan in chemical structure, but resembles bacterial peptidoglycan in function and physical structure. Pseudopeptidoglycan, in general, is only present in a few methanogenic archaea. The basic components are N-acetylglucosamine and N-acetyltalosaminuronic acid (bacterial peptidoglycan containing N-acetylmuramic acid instead), which are linked by β-1,3-glycosidic bonds.[Albers, Sonja; Eichler, Jerry; Aebi, Markus (2015), Varki, Ajit; Cummings, Richard D.; Esko, Jeffrey D.; Stanley, Pamela (eds.), “Archaea”, Essentials of Glycobiology (3rd ed.), Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press, doi:10.1101/glycobiology.3e.022 (inactive 1 August 2023), PMID 28876866, retrieved 2021-04-19] Lysozyme, a host defense mechanism present in human secretions (e.g. saliva and tears) breaks β-1,4-glycosidic bonds to degrade peptidoglycan. However, because pseudopeptidoglycan has β-1,3-glycosidic bonds, lysozyme is ineffective. It was thought from these large differences in cell wall chemistry that archaeal cell walls and bacterial cell walls have not evolved from a common ancestor but are only the result of a convergent evolution,[Visweswaran, Ganesh Ram R.; Dijkstra, Bauke W.; Kok, Jan (2011). “Murein and pseudomurein cell wall binding domains of bacteria and archaea—a comparative view”. Applied Microbiology and Biotechnology. 92 (5): 921–928. doi:10.1007/s00253-011-3637-0. ISSN 0175-7598. PMC 3210951. PMID 22012341.] but recent structural work has revealed deeper homology.[Subedi, Bishwa P; Martin, William F; Carbone, Vincenzo; Duin, Eduardus C; Cronin, Bryan; Sauter, Julia; Schofield, Linley R; Sutherland-Smith, Andrew J; Ronimus, Ron S (7 September 2021). “Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry”. FEMS Microbes. 2: xtab012. doi:10.1093/femsmc/xtab012. PMC 10117817. PMID 37334239.] No archaeal enzymes are known that cleave the β-1,3-glycosidic bonds in pseudopeptidoglycan, but it can be degraded by pseudomurein endoisopeptidase encoded by two prophages.[Visweswaran, Ganesh Ram R.; Dijkstra, Bauke W.; Kok, Jan (2010). “Two Major Archaeal Pseudomurein Endoisopeptidases: PeiW and PeiP”. Archaea. 2010: 480492. doi:10.1155/2010/480492. PMC 2989375. PMID 21113291.] The pseudomurein endoisopeptidases function by cleaving the peptide links between adjacent pseudopeptidoglycan strands. Pseudopeptidoglycan is composed of two sugars, N-acetylglucosamine and N-acetyltalosaminuronic acid. These sugars are made of different amino acids, and the peptide cross-links within pseudopeptidoglycan are formed with different amino acids. The peptide bond is formed between the lysine of a N-acetyltalosaminuronic acid and a glutamine of a parallel N-acetyltalosaminuronic acid.[Slonczewski, Joan, Watkins, John, Foster.; Slonczewski, Joan (2009). Microbiology: An Evolving Science.[better source needed]] Pseudopeptidoglycan, like peptidoglycan in bacteria, forms a mesh-like layer outside of the plasma membrane of the archaea. Only a few methanogenic archaea have cell walls composed of pseudopeptidoglycan. This component functions much like peptidoglycan in a bacterial cell.[“Peptidoglycan vs Pseudopeptidoglycan | Easy Biology Class”. www.easybiologyclass.com. 2017-05-19. Retrieved 2021-05-06.] Pseudopeptidoglycan is used by the archaeal cell to determine its shape and provide structure to the cell. It is also used to protect the cell from undesired molecules or anything harmful in its environment. PPG is produced by enzymes of two gene clusters. Recent work on the peptide ligases show, surprisingly, a common origin with murein synthesis. The pathway is now known to include the orthologous-to-bacteria CarB, MurC/D (peptide ligase), MurG, MraY, UppP, UppS, and flippase presumably performing an analogous function, and two novel but conserved transmembrane proteins. GlmM and GlmU, which produce UDP-GlcNAc in bacteria, are also present with phosphoglucomutase (PGM). Half of the species also have MurT and GatD, known to perform cell wall modifications in bacteria. No orthologous cross-linking enzymes have been identified. Notably, “formation of the disaccharide moiety of the glycopeptide monomer occurs before the transfer to membrane protein by MraY”, as opposed to after in bacteria. Further work would be needed to connect these information into a coherent pathway.[Subedi, Bishwa P; Martin, William F; Carbone, Vincenzo; Duin, Eduardus C; Cronin, Bryan; Sauter, Julia; Schofield, Linley R; Sutherland-Smith, Andrew J; Ronimus, Ron S (7 September 2021). “Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry”. FEMS Microbes. 2: xtab012. doi:10.1093/femsmc/xtab012. PMC 10117817. PMID 37334239.] Lysozyme is a natural defense mechanism in humans that has the ability to break down peptidoglycan in bacterial cells. It degrades the peptidoglycan by targeting the β-1,4-glycosidic bonds that connect the alternating amino sugars in which it is composed of.[Primo, Emiliano D.; Otero, Lisandro H.; Ruiz, Francisco; Klinke, Sebastián; Giordano, Walter (2018). “The disruptive effect of lysozyme on the bacterial cell wall explored by an in-silico structural outlook”. Biochemistry and Molecular Biology Education. 46 (1): 83–90. doi:10.1002/bmb.21092. ISSN 1539-3429. PMID 29131507.] This degradation of the glycosidic bonds within peptidoglycan cause the sugars to separate and inhibit the structural integrity of the peptidoglycan and the bacteria. Pseudopeptidoglycan, however, is composed of a different acidic amino sugar, which is N-acetyltalosaminuronic acid. This difference is the reason that it has β-1,3-glycosidic bonds (as opposed to the β-1,4-glycosidic bonds in bacteria).[Albers, Sonja; Eichler, Jerry; Aebi, Markus (2015), Varki, Ajit; Cummings, Richard D.; Esko, Jeffrey D.; Stanley, Pamela (eds.), “Archaea”, Essentials of Glycobiology (3rd ed.), Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press, doi:10.1101/glycobiology.3e.022 (inactive 1 August 2023), PMID 28876866, retrieved 2021-04-19] Lysozymes targets the linkage in peptidoglycan, and without that, becomes ineffective against pseudopeptidoglycan. Penicillin is a group of antibiotics that have been effective against many bacterial infections. It attacks bacteria by targeting and inhibiting the transpeptidase that catalyzes the cross-linking of the amino sugars in peptidoglycan.[Yocum, R. R.; Rasmussen, J. R.; Strominger, J. L. (1980-05-10). “The mechanism of action of penicillin. Penicillin acylates the active site of Bacillus stearothermophilus D-alanine carboxypeptidase”. The Journal of Biological Chemistry. 255 (9): 3977–3986. doi:10.1016/S0021-9258(19)85621-1. ISSN 0021-9258. PMID 7372662.] However, peptidoglycan contains different amino sugars, and therefore, a different catalysis enzyme is used. The different amino acids cause antibiotics, that target cell walls like penicillin, to be ineffective against pseudopeptidoglycan.[Slonczewski, Joan, Watkins, John, Foster.; Slonczewski, Joan (2009). Microbiology: An Evolving Science.[better source needed]] PPG is found in the archaeal orders of Methanobacteriales and Methanopyrales.[Subedi, Bishwa P; Martin, William F; Carbone, Vincenzo; Duin, Eduardus C; Cronin, Bryan; Sauter, Julia; Schofield, Linley R; Sutherland-Smith, Andrew J; Ronimus, Ron S (7 September 2021). “Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry”. FEMS Microbes. 2: xtab012. doi:10.1093/femsmc/xtab012. PMC 10117817. PMID 37334239.] Some genera under these orders are: Methanobacterium, Methanobrevibacter, Methanothermobacter, Methanothermus, Methanosphaera, Methanopyrus. See also: Cell wall and Methanochondroitin
- Methanochondroitin, which is similar to eukaryotic connective tissue chondroitin, is composed of a repeating trimer of uronic acid and two GalNAc residues. Yet unlike chondroitin, methanochondroitin is not sulfated. A pathway of methanochondroitin biosynthesis has been proposed based on activated precursors in Methanosarcina barkeri cell extracts. Methanosarcina species can further modify the methanochondroitin condition largely through the addition of glucose and galactose acids. [Meyer BH, Albers SV, Eichler J, et al. Archaea. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology [Internet]. 4th edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022. Chapter 22. Available from: https://www.ncbi.nlm.nih.gov/books/NBK579933/ doi: 10.1101/glycobiology.4e.22]
Recognition by immune system
Peptidoglycan recognition is an evolutionarily conserved process. The overall structure is similar between bacterial species, but various modifications can increase the diversity. These include modifications of the length of sugar polymers, modifications in the sugar structures, variations in cross-linking or substitutions of amino acids (primarily at the third position). The aim of these modifications is to alter the properties of the cell wall, which plays a vital role in pathogenesis.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
- Bersch KL, DeMeester KE, Zagani R, Chen S, Wodzanowski KA, Liu S, et al. (April 2021). “Bacterial Peptidoglycan Fragments Differentially Regulate Innate Immune Signaling”. ACS Central Science. 7 (4): 688–696. doi:10.1021/acscentsci.1c00200. PMC 8155477. PMID 34056099.
Peptidoglycans can be degraded by several enzymes (lysozyme, glucosaminidase, endopeptidase…), producing immunostimulatory fragments (sometimes called muropeptides) that are critical for mediating host-pathogen interactions. These include MDP (muramyl dipeptide), NAG (N-acetylglucosamine) or iE-DAP (γ-d-glutamyl-meso-diaminopimelic acid).
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
- Bersch KL, DeMeester KE, Zagani R, Chen S, Wodzanowski KA, Liu S, et al. (April 2021). “Bacterial Peptidoglycan Fragments Differentially Regulate Innate Immune Signaling”. ACS Central Science. 7 (4): 688–696. doi:10.1021/acscentsci.1c00200. PMC 8155477. PMID 34056099.
Peptidoglycan from intestinal bacteria (both pathogens and commensals) crosses the intestinal barrier even under physiological conditions. Mechanisms through which peptidoglycan or its fragments enter the host cells can be direct (carrier-independent) or indirect (carrier-dependent), and they are either bacteria-mediated (secretion systems, membrane vesicles) or host cell-mediated (receptor-mediated, peptide transporters).
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
Bacterial secretion systems are protein complexes used for the delivery of virulence factors across the bacterial cell envelope to the exterior environment.
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
Intracellular bacterial pathogens invade eukaryotic cells (which may lead to the formation of phagolysosomes and/or autophagy activation), or bacteria may be engulfed by phagocytes (macrophages, monocytes, neutrophils…). The bacteria-containing phagosome may then fuse with endosomes and lysosomes, leading to degradation of bacteria and generation of polymeric peptidoglycan fragments and muropeptides.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
Receptors
Innate immune system senses intact peptidoglycan and peptidoglycan fragments using numerous PRRs (pattern recognition receptors) that are secreted, expressed intracellularly or expressed on the cell surface.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
Peptidoglycan recognition proteins
PGLYRPs are conserved from insects to mammals. Mammals produce four secreted soluble peptidoglycan recognition proteins (PGLYRP-1, PGLYRP-2, PGLYRP-3 and PGLYRP-4) that recognize muramyl pentapeptide or tetrapeptide. They can also bind to LPS and other molecules by using binding sites outside of the peptidoglycan-binding groove. After recognition of peptidoglycan, PGLYRPs activate polyphenol oxidase (PPO) molecules, Toll, or immune deficiency (IMD) signalling pathways. That leads to production of antimicrobial peptides (AMPs).
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
Each of the mammalian PGLYRPs display unique tissue expression patterns. PGLYRP-1 is mainly expressed in the granules of neutrophils and eosinophils.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
PGLYRP-3 and 4 are expressed by several tissues such as skin, sweat glands, eyes or the intestinal tract. PGLYRP-1, 3 and 4 form disulphide-linked homodimers and heterodimers essential for their bactericidal activity.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
Their binding to bacterial cell wall peptidoglycans can induce bacterial cell death by interaction with various bacterial transcriptional regulatory proteins. PGLYRPs are likely to assist in bacterial killing by cooperating with other PRRs to enhance recognition of bacteria by phagocytes.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
PGLYRP-2 is primarily expressed by the liver and secreted into the circulation.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
Also, its expression can be induced in skin keratinocytes, oral and intestinal epithelial cells.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
In contrast with the other PGLYRPs, PGLYRP-2 has no direct bactericidal activity. It possesses peptidoglycan amidase activity, it hydrolyses the lactyl-amide bond between the MurNAc and the first amino acid of the stem peptide of peptidoglycan.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
It is proposed, that the function of PGLYRP-2 is to prevent over-activation of the immune system and inflammation-induced tissue damage in response to NOD2 ligands (see below), as these muropeptides can no longer be recognized by NOD2 upon separation of the peptide component from MurNAc.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
Growing evidence suggests that peptidoglycan recognition protein family members play a dominant role in the tolerance of intestinal epithelial cells toward the commensal microbiota.
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
- Liang Y, Yang L, Wang Y, Tang T, Liu F, Zhang F (December 2022). “Peptidoglycan recognition protein SC (PGRP-SC) shapes gut microbiota richness, diversity and composition by modulating immunity in the house fly Musca domestica”. Insect Molecular Biology. 32 (2): 200–212. doi:10.1111/imb.12824. PMID 36522831. S2CID 254807823.
It has been demonstrated that expression of PGLYRP-2 and 4 can influence the composition of the intestinal microbiota.
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
Recently, it has been discovered, that PGLYRPs (and also NOD-like receptors and peptidoglycan transporters) are highly expressed in the developing mouse brain. PGLYRP-2 and is highly expressed in neurons of several brain regions including the prefrontal cortex, hippocampus, and cerebellum, thus indicating potential direct effects of peptidoglycan on neurons. PGLYRP-2 is highly expressed also in the cerebral cortex of young children, but not in most adult cortical tissues. PGLYRP-1 is also expressed in the brain and continues to be expressed into adulthood.
- Gonzalez-Santana A, Diaz Heijtz R (August 2020). “Bacterial Peptidoglycans from Microbiota in Neurodevelopment and Behavior” (PDF). Trends in Molecular Medicine. 26 (8): 729–743. doi:10.1016/j.molmed.2020.05.003. PMID 32507655. S2CID 219539658.
Peptidoglycan recognition proteins (PGRPs) are a group of highly conserved pattern recognition receptors with at least one peptidoglycan recognition domain capable of recognizing the peptidoglycan component of the cell wall of bacteria. They are present in insects, mollusks, echinoderms and chordates. The mechanism of action of PGRPs varies between taxa. In insects, PGRPs kill bacteria indirectly by activating one of four unique effector pathways: prophenoloxidase cascade, Toll pathway, IMD pathway, and induction of phagocytosis. In mammals, PGRPs either kill bacteria directly by interacting with their cell wall or outer membrane, or hydrolyze peptidoglycan.[ Royet, Julien; Gupta, Dipika; Dziarski, Roman (11 November 2011). “Peptidoglycan recognition proteins: modulators of the microbiome and inflammation”. Nature Reviews Immunology. 11 (12): 837–51. doi:10.1038/nri3089. PMID 22076558. S2CID 5266193.][Royet, Julien; Dziarski, Roman (April 2007). “Peptidoglycan recognition proteins: pleiotropic sensors and effectors of antimicrobial defences”. Nature Reviews Microbiology. 5 (4): 264–277. doi:10.1038/nrmicro1620. ISSN 1740-1526. PMID 17363965. S2CID 39569790.][Dziarski, Roman; Royet, Julien; Gupta, Dipika (2016), “Peptidoglycan Recognition Proteins and Lysozyme”, Encyclopedia of Immunobiology, Elsevier, pp. 389–403, doi:10.1016/b978-0-12-374279-7.02022-1, ISBN 978-0-08-092152-5, retrieved 2020-10-22][Dziarski, Roman; Gupta, Dipika (2006). “The peptidoglycan recognition proteins (PGRPs)”. Genome Biology. 7 (8): 232. doi:10.1186/gb-2006-7-8-232. PMC 1779587. PMID 16930467.] They also modulate inflammation and microbiome and interact with host receptors. The first PGRP was discovered in 1996 by Masaaki Ashida and coworkers, who purified a 19 kDa protein present in the hemolymph and cuticle of a silkworm (Bombyx mori), and named it Peptidoglycan Recognition Protein, because it specifically bound peptidoglycan and activated the prophenoloxidase cascade.[Yoshida, Hideya; Kinoshita, Kuninori; Ashida, Masaaki (1996-06-07). “Purification of a Peptidoglycan Recognition Protein from Hemolymph of the Silkworm, Bombyx mori”. Journal of Biological Chemistry. 271 (23): 13854–13860. doi:10.1074/jbc.271.23.13854. ISSN 0021-9258. PMID 8662762. S2CID 20831557.] In 1998 Håkan Steiner and coworkers, using a differential display screen, identified and cloned a PGRP ortholog in a moth (Trichoplusia ni) and then discovered and cloned mouse and human PGRP orthologs,[Kang, D.; Liu, G.; Lundstrom, A.; Gelius, E.; Steiner, H. (1998-08-18). “A peptidoglycan recognition protein in innate immunity conserved from insects to humans”. Proceedings of the National Academy of Sciences. 95 (17): 10078–10082. Bibcode:1998PNAS…9510078K. doi:10.1073/pnas.95.17.10078. ISSN 0027-8424. PMC 21464. PMID 9707603.] thus showing that PGRPs are highly conserved from insects to mammals. Also in 1998, Sergei Kiselev and coworkers independently discovered and cloned a protein from a mouse adenocarcinoma with the same sequence as PGRP, which they named Tag7.[Kiselev, Sergei L.; Kustikova, Olga S.; Korobko, Elena V.; Prokhortchouk, Egor B.; Kabishev, Andrei A.; Lukanidin, Evgenii M.; Georgiev, Georgii P. (1998-07-17). “Molecular Cloning and Characterization of the Mouse tag7 Gene Encoding a Novel Cytokine”. Journal of Biological Chemistry. 273 (29): 18633–18639. doi:10.1074/jbc.273.29.18633. ISSN 0021-9258. PMID 9660837. S2CID 11417742.] In 1999 Masanori Ochiai and Masaaki Ashida cloned the silkworm (B. mori) PGRP.[Ochiai, Masanori; Ashida, Masaaki (1999-04-23). “A Pattern Recognition Protein for Peptidoglycan: CLONING THE cDNA AND THE GENE OF THE SILKWORM, BOMBYX MORI”. Journal of Biological Chemistry. 274 (17): 11854–11858. doi:10.1074/jbc.274.17.11854. ISSN 0021-9258. PMID 10207004. S2CID 38022527.]…Genetic PGLYRP variants or changed expression of PGRPs are associated with several diseases. Patients with inflammatory bowel disease (IBD), which includes Crohn’s disease and ulcerative colitis, have significantly more frequent missense variants in all four PGLYRP genes than healthy controls.[Zulfiqar, Fareeha; Hozo, Iztok; Rangarajan, Sneha; Mariuzza, Roy A.; Dziarski, Roman; Gupta, Dipika (2013). “Genetic Association of Peptidoglycan Recognition Protein Variants with Inflammatory Bowel Disease”. PLOS ONE. 8 (6): e67393. Bibcode:2013PLoSO…867393Z. doi:10.1371/journal.pone.0067393. ISSN 1932-6203. PMC 3686734. PMID 23840689.] These results suggest that PGRPs protect humans from these inflammatory diseases, and that mutations in PGLYRP genes are among the genetic factors predisposing to these diseases. PGLYRP1 variants are also associated with increased fetal hemoglobin in sickle cell disease,[Nkya, Siana; Mwita, Liberata; Mgaya, Josephine; Kumburu, Happiness; van Zwetselaar, Marco; Menzel, Stephan; Mazandu, Gaston Kuzamunu; Sangeda, Raphael; Chimusa, Emile; Makani, Julie (5 June 2020). “Identifying genetic variants and pathways associated with extreme levels of fetal hemoglobin in sickle cell disease in Tanzania”. BMC Medical Genetics. 21 (1): 125. doi:10.1186/s12881-020-01059-1. ISSN 1471-2350. PMC 7275552. PMID 32503527.] PGLYRP2 variants are associated with esophageal squamous cell carcinoma,[Ng, David; Hu, Nan; Hu, Ying; Wang, Chaoyu; Giffen, Carol; Tang, Ze-Zhong; Han, Xiao-You; Yang, Howard H.; Lee, Maxwell P.; Goldstein, Alisa M.; Taylor, Philip R. (2008-10-01). “Replication of a genome-wide case-control study of esophageal squamous cell carcinoma”. International Journal of Cancer. 123 (7): 1610–1615. doi:10.1002/ijc.23682. ISSN 1097-0215. PMC 2552411. PMID 18649358.] PGLYRP2, PGLYRP3, and PGLYRP4 variants are associated with Parkinson’s disease,[Goldman, Samuel M.; Kamel, Freya; Ross, G. Webster; Jewell, Sarah A.; Marras, Connie; Hoppin, Jane A.; Umbach, David M.; Bhudhikanok, Grace S.; Meng, Cheryl; Korell, Monica; Comyns, Kathleen (August 2014). “Peptidoglycan recognition protein genes and risk of Parkinson’s disease”. Movement Disorders. 29 (9): 1171–1180. doi:10.1002/mds.25895. ISSN 1531-8257. PMC 4777298. PMID 24838182.] PGLYRP3 and PGLYRP4 variants are associated with psoriasis[Sun, Chao; Mathur, Punam; Dupuis, Josée; Tizard, Rich; Ticho, Barry; Crowell, Tom; Gardner, Humphrey; Bowcock, Anne M.; Carulli, John (March 2006). “Peptidoglycan recognition proteins Pglyrp3 and Pglyrp4 are encoded from the epidermal differentiation complex and are candidate genes for the Psors4 locus on chromosome 1q21”. Human Genetics. 119 (1–2): 113–125. doi:10.1007/s00439-005-0115-8. ISSN 0340-6717. PMID 16362825. S2CID 31486449.][Kainu, Kati; Kivinen, Katja; Zucchelli, Marco; Suomela, Sari; Kere, Juha; Inerot, Annica; Baker, Barbara S.; Powles, Anne V.; Fry, Lionel; Samuelsson, Lena; Saarialho-Kere, Ulpu (February 2009). “Association of psoriasis to PGLYRP and SPRR genes at PSORS4 locus on 1q shows heterogeneity between Finnish, Swedish and Irish families”. Experimental Dermatology. 18 (2): 109–115. doi:10.1111/j.1600-0625.2008.00769.x. ISSN 1600-0625. PMID 18643845. S2CID 5771478.] and composition of airway microbiome,[Igartua, Catherine; Davenport, Emily R.; Gilad, Yoav; Nicolae, Dan L.; Pinto, Jayant; Ober, Carole (1 February 2017). “Host genetic variation in mucosal immunity pathways influences the upper airway microbiome”. Microbiome. 5 (1): 16. doi:10.1186/s40168-016-0227-5. ISSN 2049-2618. PMC 5286564. PMID 28143570.] and PGLYRP4 variants are associated with ovarian cancer.[Zhang, Lei; Luo, Min; Yang, Hongying; Zhu, Shaoyan; Cheng, Xianliang; Qing, Chen (2019-02-20). “Next-generation sequencing-based genomic profiling analysis reveals novel mutations for clinical diagnosis in Chinese primary epithelial ovarian cancer patients”. Journal of Ovarian Research. 12 (1): 19. doi:10.1186/s13048-019-0494-4. ISSN 1757-2215. PMC 6381667. PMID 30786925.] Several diseases are associated with increased expression of PGLYRP1, including: atherosclerosis,[Rohatgi, Anand; Ayers, Colby R.; Khera, Amit; McGuire, Darren K.; Das, Sandeep R.; Matulevicius, Susan; Timaran, Carlos H.; Rosero, Eric B.; de Lemos, James A. (April 2009). “The association between peptidoglycan recognition protein-1 and coronary and peripheral atherosclerosis: Observations from the Dallas Heart Study”. Atherosclerosis. 203 (2): 569–575. doi:10.1016/j.atherosclerosis.2008.07.015. ISSN 1879-1484. PMID 18774573.][Brownell, Nicholas K.; Khera, Amit; de Lemos, James A.; Ayers, Colby R.; Rohatgi, Anand (17 May 2016). “Association Between Peptidoglycan Recognition Protein-1 and Incident Atherosclerotic Cardiovascular Disease Events: The Dallas Heart Study”. Journal of the American College of Cardiology. 67 (19): 2310–2312. doi:10.1016/j.jacc.2016.02.063. ISSN 1558-3597. PMID 27173041.] myocardial infarction, heart failure,[Klimczak-Tomaniak, Dominika; Bouwens, Elke; Schuurman, Anne-Sophie; Akkerhuis, K. Martijn; Constantinescu, Alina; Brugts, Jasper; Westenbrink, B. Daan; van Ramshorst, Jan; Germans, Tjeerd; Pączek, Leszek; Umans, Victor (June 2020). “Temporal patterns of macrophage- and neutrophil-related markers are associated with clinical outcome in heart failure patients”. ESC Heart Failure. 7 (3): 1190–1200. doi:10.1002/ehf2.12678. ISSN 2055-5822. PMC 7261550. PMID 32196993.] sepsis,[Zhang, Junli; Cheng, Yuelei; Duan, Minmin; Qi, Nannan; Liu, Jian (May 2017). “Unveiling differentially expressed genes upon regulation of transcription factors in sepsis”. 3 Biotech. 7 (1): 46. doi:10.1007/s13205-017-0713-x. ISSN 2190-572X. PMC 5428098. PMID 28444588.] pulmonary fibrosis,[Molyneaux, Philip L.; Willis-Owen, Saffron A. G.; Cox, Michael J.; James, Phillip; Cowman, Steven; Loebinger, Michael; Blanchard, Andrew; Edwards, Lindsay M.; Stock, Carmel; Daccord, Cécile; Renzoni, Elisabetta A. (15 June 2017). “Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis”. American Journal of Respiratory and Critical Care Medicine. 195 (12): 1640–1650. doi:10.1164/rccm.201607-1408OC. ISSN 1535-4970. PMC 5476909. PMID 28085486.] asthma,[Kasaian, M. T.; Lee, J.; Brennan, A.; Danto, S. I.; Black, K. E.; Fitz, L.; Dixon, A. E. (July 2018). “Proteomic analysis of serum and sputum analytes distinguishes controlled and poorly controlled asthmatics”. Clinical and Experimental Allergy. 48 (7): 814–824. doi:10.1111/cea.13151. ISSN 1365-2222. PMID 29665127. S2CID 4938216.] chronic kidney disease,[Nylund, Karita M.; Ruokonen, Hellevi; Sorsa, Timo; Heikkinen, Anna Maria; Meurman, Jukka H.; Ortiz, Fernanda; Tervahartiala, Taina; Furuholm, Jussi; Bostanci, Nagihan (January 2018). “Association of the salivary triggering receptor expressed on myeloid cells/its ligand peptidoglycan recognition protein 1 axis with oral inflammation in kidney disease”. Journal of Periodontology. 89 (1): 117–129. doi:10.1902/jop.2017.170218. ISSN 1943-3670. PMID 28846062. S2CID 21830535.] rheumatoid arthritis,[Luo, Qing; Li, Xue; Zhang, Lu; Yao, Fangyi; Deng, Zhen; Qing, Cheng; Su, Rigu; Xu, Jianqing; Guo, Yang; Huang, Zikun; Li, Junming (January 2019). “Serum PGLYRP‑1 is a highly discriminatory biomarker for the diagnosis of rheumatoid arthritis”. Molecular Medicine Reports. 19 (1): 589–594. doi:10.3892/mmr.2018.9632. ISSN 1791-3004. PMID 30431075.] gingival inflammation,[Silbereisen, A.; Hallak, A. K.; Nascimento, G. G.; Sorsa, T.; Belibasakis, G. N.; Lopez, R.; Bostanci, N. (October 2019). “Regulation of PGLYRP1 and TREM-1 during Progression and Resolution of Gingival Inflammation”. JDR Clinical and Translational Research. 4 (4): 352–359. doi:10.1177/2380084419844937. ISSN 2380-0852. PMID 31013451. S2CID 129941967.][Raivisto, T.; Heikkinen, A. M.; Silbereisen, A.; Kovanen, L.; Ruokonen, H.; Tervahartiala, T.; Haukka, J.; Sorsa, T.; Bostanci, N. (October 2020). “Regulation of Salivary Peptidoglycan Recognition Protein 1 in Adolescents”. JDR Clinical and Translational Research. 5 (4): 332–341. doi:10.1177/2380084419894287. ISSN 2380-0852. PMID 31860804. S2CID 209434091.][Yucel, Zeynep Pinar Keles; Silbereisen, Angelika; Emingil, Gulnur; Tokgoz, Yavuz; Kose, Timur; Sorsa, Timo; Tsilingaridis, Georgios; Bostanci, Nagihan (October 2020). “Salivary biomarkers in the context of gingival inflammation in children with cystic fibrosis”. Journal of Periodontology. 91 (10): 1339–1347. doi:10.1002/JPER.19-0415. hdl:10138/327022. ISSN 1943-3670. PMID 32100289. S2CID 211523360.][Karsiyaka Hendek, Meltem; Kisa, Ucler; Olgun, Ebru (January 2020). “The effect of smoking on gingival crevicular fluid peptidoglycan recognition protein-1 level following initial periodontal therapy in chronic periodontitis”. Oral Diseases. 26 (1): 166–172. doi:10.1111/odi.13207. ISSN 1601-0825. PMID 31587460. S2CID 203850763.][Teixeira, Mayla K. S.; Lira-Junior, Ronaldo; Lourenço, Eduardo José Veras; Telles, Daniel Moraes; Boström, Elisabeth A.; Figueredo, Carlos Marcelo; Bostanci, Nagihan (May 2020). “The modulation of the TREM-1/PGLYRP1/MMP-8 axis in peri-implant diseases”. Clinical Oral Investigations. 24 (5): 1837–1844. doi:10.1007/s00784-019-03047-z. ISSN 1436-3771. PMID 31444693. S2CID 201283050.] osteoarthritis,[Yang, Zhanyu; Ni, Jiangdong; Kuang, Letian; Gao, Yongquan; Tao, Shibin (2020-09-11). “Identification of genes and pathways associated with subchondral bone in osteoarthritis via bioinformatic analysis”. Medicine. 99 (37): e22142. doi:10.1097/MD.0000000000022142. ISSN 1536-5964. PMC 7489699. PMID 32925767.] cardiovascular events and death in kidney transplant patients,[Ortiz, Fernanda; Nylund, Karita M.; Ruokonen, Hellevi; Meurman, Jukka H.; Furuholm, Jussi; Bostanci, Nagihan; Sorsa, Timo (2020-08-04). “Salivary Biomarkers of Oral Inflammation Are Associated With Cardiovascular Events and Death Among Kidney Transplant Patients”. Transplantation Proceedings. 52 (10): 3231–3235. doi:10.1016/j.transproceed.2020.07.007. ISSN 1873-2623. PMID 32768288. S2CID 225451024.] alopecia,[Glickman, Jacob W.; Dubin, Celina; Renert-Yuval, Yael; Dahabreh, Dante; Kimmel, Grace W.; Auyeung, Kelsey; Estrada, Yeriel D.; Singer, Giselle; Krueger, James G.; Pavel, Ana B.; Guttman-Yassky, Emma (2020-05-04). “Cross-sectional study of blood biomarkers of patients with moderate to severe alopecia areata reveals systemic immune and cardiovascular biomarker dysregulation”. Journal of the American Academy of Dermatology. 84 (2): 370–380. doi:10.1016/j.jaad.2020.04.138. ISSN 1097-6787. PMID 32376430. S2CID 218532915.] type I diabetes,[Yang, Shuting; Cao, Chuqing; Xie, Zhiguo; Zhou, Zhiguang (March 2020). “Analysis of potential hub genes involved in the pathogenesis of Chinese type 1 diabetic patients”. Annals of Translational Medicine. 8 (6): 295. doi:10.21037/atm.2020.02.171. ISSN 2305-5839. PMC 7186604. PMID 32355739.] infectious complications in hemodialysis,[Arenius, Ilona; Ruokonen, Hellevi; Ortiz, Fernanda; Furuholm, Jussi; Välimaa, Hannamari; Bostanci, Nagihan; Eskola, Maija; Maria Heikkinen, Anna; Meurman, Jukka H.; Sorsa, Timo; Nylund, Karita (July 2020). “The relationship between oral diseases and infectious complications in patients under dialysis”. Oral Diseases. 26 (5): 1045–1052. doi:10.1111/odi.13296. hdl:10138/325947. ISSN 1601-0825. PMID 32026534. S2CID 211045697.] and thrombosis,[Guo, Chao; Li, Zhenling (2019-12-05). “Bioinformatics Analysis of Key Genes and Pathways Associated with Thrombosis in Essential Thrombocythemia”. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 25: 9262–9271. doi:10.12659/MSM.918719. ISSN 1643-3750. PMC 6911306. PMID 31801935.] consistent with pro-inflammatory effects of PGLYRP1. Lower expression of PGLYRP1 was found in endometriosis.[Grande, Giuseppe; Vincenzoni, Federica; Milardi, Domenico; Pompa, Giuseppina; Ricciardi, Domenico; Fruscella, Erika; Mancini, Francesca; Pontecorvi, Alfredo; Castagnola, Massimo; Marana, Riccardo (2017). “Cervical mucus proteome in endometriosis”. Clinical Proteomics. 14: 7. doi:10.1186/s12014-017-9142-4. ISSN 1542-6416. PMC 5290661. PMID 28174513.] Decreased expression of PGLYRP2 is associated with HIV-associated tuberculosis,[Achkar, Jacqueline M.; Cortes, Laetitia; Croteau, Pascal; Yanofsky, Corey; Mentinova, Marija; Rajotte, Isabelle; Schirm, Michael; Zhou, Yiyong; Junqueira-Kipnis, Ana Paula; Kasprowicz, Victoria O.; Larsen, Michelle (September 2015). “Host Protein Biomarkers Identify Active Tuberculosis in HIV Uninfected and Co-infected Individuals”. EBioMedicine. 2 (9): 1160–1168. doi:10.1016/j.ebiom.2015.07.039. ISSN 2352-3964. PMC 4588417. PMID 26501113.] Lyme disease,[Zhou, Yong; Qin, Shizhen; Sun, Mingjuan; Tang, Li; Yan, Xiaowei; Kim, Taek-Kyun; Caballero, Juan; Glusman, Gustavo; Brunkow, Mary E.; Soloski, Mark J.; Rebman, Alison W. (3 January 2020). “Measurement of Organ-Specific and Acute-Phase Blood Protein Levels in Early Lyme Disease”. Journal of Proteome Research. 19 (1): 346–359. doi:10.1021/acs.jproteome.9b00569. ISSN 1535-3907. PMC 7981273. PMID 31618575.] hepatocellular carcinoma,[Yang, Zongyi; Feng, Jia; Xiao, Li; Chen, Xi; Yao, Yuanfei; Li, Yiqun; Tang, Yu; Zhang, Shuai; Lu, Min; Qian, Yu; Wu, Hongjin (May 2020). “Tumor-Derived Peptidoglycan Recognition Protein 2 Predicts Survival and Antitumor Immune Responses in Hepatocellular Carcinoma”. Hepatology. 71 (5): 1626–1642. doi:10.1002/hep.30924. ISSN 1527-3350. PMC 7318564. PMID 31479523.] and myocardial infarction.[Das, Apabrita Ayan; Choudhury, Kamalika Roy; Jagadeeshaprasad, M. G.; Kulkarni, Mahesh J.; Mondal, Prakash Chandra; Bandyopadhyay, Arun (2020-06-30). “Proteomic analysis detects deregulated reverse cholesterol transport in human subjects with ST-segment elevation myocardial infarction”. Journal of Proteomics. 222: 103796. doi:10.1016/j.jprot.2020.103796. ISSN 1876-7737. PMID 32376501. S2CID 218532507.] A silkworm larvae plasma (SLP) test to detect peptidoglycan, based on activation of the prophenoloxidase cascade by PGRP in the hemolymph of the silkworm, Bombyx mori, is available.[suchiya, M.; Asahi, N.; Suzuoki, F.; Ashida, M.; Matsuura, S. (September 1996). “Detection of peptidoglycan and beta-glucan with silkworm larvae plasma test”. FEMS Immunology and Medical Microbiology. 15 (2–3): 129–134. doi:10.1111/j.1574-695X.1996.tb00063.x. ISSN 0928-8244. PMID 8880138.][Kobayashi, T.; Tani, T.; Yokota, T.; Kodama, M. (May 2000). “Detection of peptidoglycan in human plasma using the silkworm larvae plasma test”. FEMS Immunology and Medical Microbiology. 28 (1): 49–53. doi:10.1111/j.1574-695X.2000.tb01456.x. ISSN 0928-8244. PMID 10767607.]
NOD-like receptors
Probably the most well-known receptors of peptidoglycan are the NOD-like receptors (NLRs), mainly NOD1 and NOD2. The NOD1 receptor is activated after iE-DAP (γ-d-glutamyl-meso-diaminopimelic acid) binding, while NOD2 recognizes MDP (muramyl dipeptide), by their LRR domains. Activation leads to self-oligomerization, resulting in activation of two signalling cascades. One triggers activation of NF-κB (through RIP2, TAK1 and IKK), second leads to MAPK signalling cascade. Activation of these pathways induces production of inflammatory cytokines and chemokines.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
- Murphy K (2017). Janeway’s immunobiology. Casey Weaver, Charles Janeway (9th ed.). New York. pp. 45, 96–98. ISBN 978-0-8153-4505-3. OCLC 933586700.
NOD1 is expressed by diverse cell types, including myeloid phagocytes, epithelial cells and neurons.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
- Gonzalez-Santana A, Diaz Heijtz R (August 2020). “Bacterial Peptidoglycans from Microbiota in Neurodevelopment and Behavior” (PDF). Trends in Molecular Medicine. 26 (8): 729–743. doi:10.1016/j.molmed.2020.05.003. PMID 32507655. S2CID 219539658.
NOD2 is expressed in monocytes and macrophages, epithelial intestinal cells, Paneth cells, dendritic cells, osteoblasts, keratinocytes and other epithelial cell types.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
As cytosolic sensors, NOD1 and NOD2 must either detect bacteria that enter the cytosol, or peptidoglycan must be degraded to generate fragments that must be transported into the cytosol for these sensors to function.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
Recently, it was demonstrated that NLRP3 is activated by peptidoglycan, through a mechanism that is independent of NOD1 and NOD2. In macrophages, N-acetylglucosamine generated by peptidoglycan degradation was found to inhibit hexokinase activity and induce its release from the mitochondrial membrane. It promotes NLRP3 inflammasome activation through a mechanism triggered by increased mitochondrial membrane permeability.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
NLRP1 is also considered as a cytoplasmic sensor of peptidoglycan. It can sense MDP and promote IL-1 secretion through binding NOD2.[26]
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
C-type lectin receptors (CLRs)
C-type lectins are a diverse superfamily of mainly Ca2+-dependent proteins that bind a variety of carbohydrates (including the glycan skeleton of peptidoglycan), and function as innate immune receptors. CLR proteins that bind to peptidoglycan include MBL (mannose binding lectin), ficolins, Reg3A (regeneration gene family protein 3A) and PTCLec1. In mammals, they initiate the lectin-pathway of the complement cascade.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
Toll-like receptors
The role of TLRs in direct recognition of peptidoglycan is controversial. In some studies, has been reported that peptidoglycan is sensed by TLR2. But this TLR2-inducing activity could be due to cell wall lipoproteins and lipoteichoic acids that commonly co-purify with peptidoglycan. Also variation in peptidoglycan structure in bacteria from species to species may contribute to the differing results on this topic.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
- Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, Golenbock D (July 1999). “Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2”. Journal of Immunology. 163 (1): 1–5. doi:10.4049/jimmunol.163.1.1. PMID 10384090. S2CID 23630870.
As vaccine or adjuvant
Peptidoglycan is immunologically active, which can stimulate immune cells to increase the expression of cytokines and enhance antibody-dependent specific response when combined with vaccine or as adjuvant alone. MDP, which is the basic unit of peptidoglycan, was initially used as the active component of Freund’s adjuvant.
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
Freund’s adjuvant is a solution of antigen emulsified in mineral oil and used as an immunopotentiator (booster). The complete form, Freund’s Complete Adjuvant (FCA or CFA) is composed of inactivated and dried mycobacteria (usually M. tuberculosis), whereas the incomplete form (FIA or IFA) lacks the mycobacterial components (hence just the water in oil emulsion). It is named after Jules T. Freund. Freund’s complete adjuvant is effective in stimulating cell-mediated immunity and leads to potentiation of T helper cells that leads to the production of certain immunoglobulins and effector T cells. Its use in humans is forbidden by regulatory authorities, due to its toxicity. Even for animal research there are currently guidelines associated with its use, due to its painful reaction and potential for tissue damage. Injections of FCA should be subcutaneous or intraperitoneal, because intradermal injections may cause skin ulceration and necrosis; intramuscular injections may lead to temporary or permanent muscle lesion, and intravenous injections may produce pulmonary lipid embolism.[citation needed] [ontes JA, Barin JG, Talor MV, Stickel N, Schaub J, Rose NR, Čiháková D. Complete Freund’s adjuvant induces experimental autoimmune myocarditis by enhancing IL-6 production during initiation of the immune response. Immun Inflamm Dis. 2017 Jun;5(2):163-176. doi: 10.1002/iid3.155. Epub 2017 Mar 13. PMID: 28474508; PMCID: PMC5418134.]When administered to diabetes prone non-obese diabetic (NOD) mice, Freund’s complete adjuvant (FCA) prevented juvenile-onset diabetes.[Sadelain, MW; Qin, HY; Lauzon, J; Singh, B (1990), “Prevention of type I diabetes in NOD mice by adjuvant immunotherapy”, Diabetes, 39 (5): 583–589, doi:10.2337/diabetes.39.5.583, PMID 2139617][Qin, HY; Sadelain, MW; Hitchon, C; Lauzon, J; Singh, B (1993), “Complete Freund’s adjuvant-induced T cells prevent the development and adoptive transfer of diabetes in nonobese diabetic mice”, J. Immunol., 150 (5): 2072–2080, doi:10.4049/jimmunol.150.5.2072, PMID 8436836, S2CID 9779509] When combined with spleen cells, FCA was said to have reversed diabetes.[Kodama, S; Kühtreiber, W; Fujimura, S; Dale, EA; Faustman, DL (2003), “Islet Regeneration During the Reversal of Autoimmune Diabetes in NOD Mice”, Science, 302 (5648): 1223–1227, Bibcode:2003Sci…302.1223K, doi:10.1126/science.1088949, PMID 14615542, S2CID 897696] In 2006, these claims were confirmed that even without spleen cells FCA can restore insulin producing beta cells in pancreas of NOD mice.[Couzin, J. (2006), “Diabetes Studies Conflict on Power of Spleen Cells”, Science, 311 (5768): 1694, doi:10.1126/science.311.5768.1694, PMID 16556811, S2CID 10334900] Although newspapers have described the 2006 findings as confirming the earlier experiments,[Kolata, G. (March 24, 2006), “A Controversial Therapy for Diabetes Is Verified”, The New York Times, retrieved May 3, 2010] a report from NIH was released on November 23, 2006 in Science confirming the participation of spleen cells in reversing end-stage diabetes.[New data from NIH lab confirms protocol to reverse type 1 diabetes in mice, BiologyNewsNet, November 2006][Philip E. Ross, Putting Up with Self, Scientific American, November 12, 2006] FCA is known to stimulate production of tumor necrosis factor, which is thought to kill the T-cells responsible for the autoimmune destruction of the pancreatic beta cells. Still in question is whether the regrowth of functional insulin-producing cells occurs due to differentiation and proliferation of existing pancreatic stem cells, or whether the injected spleen cells re-differentiate to an insulin-producing form. Denise Faustman, whose work has been central to developing the protocol, has suggested that both mechanisms may play a role. However, in experiments to verify and examine her work, Suri reported that DNA-based evidence yielded no sign that spleen cells were needed in pancreatic islet beta cells regeneration after the FCA treatment.[Suri, A; Calderon, B; Esparza, TJ; Frederick, K; Bittner, P; Unanue, ER (2006), “Immunological Reversal of Autoimmune Diabetes Without Hematopoietic Replacement of β Cells”, Science, 311 (5768): 1778–1780, Bibcode:2006Sci…311.1778S, doi:10.1126/science.1123500, PMID 16556846, S2CID 42150301] In pancreatic islets the β-cells regenerate following Freund’s adjuvant treatment.[Huszarik, K; Wright, B; Keller, C; Nikoopour, E (2010). “Adjuvant immunotherapy increases beta cell regenerative factor Reg2 in the pancreas of diabetic mice”. J. Immunol. 185 (9): 5120–9. doi:10.4049/jimmunol.1001596. PMID 20876350.] This is related to the induction of Th17 cells by adjuvant treatment and these cells produce Interleukin-22 (IL-22). Pancreatic islets express high levels of IL-22 receptor and IL-22 has been shown to induce islet beta cell regeneration.[Hill, T; Krougly, O; Nikoopour, E; Bellemore, S (2013). “The involvement of interleukin-22 in the expression of pancreatic beta cell regenerative Reg genes”. Cell Regeneration. 2 (1): 1–11. doi:10.1186/2045-9769-2-2. PMC 4230743. PMID 25408874.] It has also been investigated in an animal model of Parkinson’s disease,[Armentero MT, Levandis G, Nappi G, Bazzini E, Blandini F (2006), “Peripheral inflammation and neuroprotection: systemic pretreatment with complete Freund’s adjuvant reduces 6-hydroxydopamine toxicity in a rodent model of Parkinson’s disease”, Neurobiol. Dis., 24 (3): 492–505, doi:10.1016/j.nbd.2006.08.016, PMID 17023164, S2CID 7076941.] or as well used in emulsion with Myelin oligodendrocyte glycoprotein (MOG), a peptide inducing Experimental autoimmune encephalomyelitis (EAE) in animal studies for efficacy testing of multiple sclerosis treatments.[Bittner, Stefan; Afzali, Ali M.; Wiendl, Heinz; Meuth, Sven G. (2014-04-15). “Myelin Oligodendrocyte Glycoprotein (MOG35-55) Induced Experimental Autoimmune Encephalomyelitis (EAE) in C57BL/6 Mice”. Journal of Visualized Experiments (86): 51275. doi:10.3791/51275. ISSN 1940-087X. PMC 4172026. PMID 24797125.] FCA is known to stimulate production of tumor necrosis factor, which is thought to kill the T-cells responsible for the autoimmune destruction of the pancreatic beta cells. Still in question is whether the regrowth of functional insulin-producing cells occurs due to differentiation and proliferation of existing pancreatic stem cells, or whether the injected spleen cells re-differentiate to an insulin-producing form. Denise Faustman, whose work has been central to developing the protocol, has suggested that both mechanisms may play a role. However, in experiments to verify and examine her work, Suri reported that DNA-based evidence yielded no sign that spleen cells were needed in pancreatic islet beta cells regeneration after the FCA treatment.[Suri, A; Calderon, B; Esparza, TJ; Frederick, K; Bittner, P; Unanue, ER (2006), “Immunological Reversal of Autoimmune Diabetes Without Hematopoietic Replacement of β Cells”, Science, 311 (5768): 1778–1780, Bibcode:2006Sci…311.1778S, doi:10.1126/science.1123500, PMID 16556846, S2CID 42150301] In pancreatic islets the β-cells regenerate following Freund’s adjuvant treatment.[Huszarik, K; Wright, B; Keller, C; Nikoopour, E (2010). “Adjuvant immunotherapy increases beta cell regenerative factor Reg2 in the pancreas of diabetic mice”. J. Immunol. 185 (9): 5120–9. doi:10.4049/jimmunol.1001596. PMID 20876350.] This is related to the induction of Th17 cells by adjuvant treatment and these cells produce Interleukin-22 (IL-22). Pancreatic islets express high levels of IL-22 receptor and IL-22 has been shown to induce islet beta cell regeneration.[Hill, T; Krougly, O; Nikoopour, E; Bellemore, S (2013). “The involvement of interleukin-22 in the expression of pancreatic beta cell regenerative Reg genes”. Cell Regeneration. 2 (1): 1–11. doi:10.1186/2045-9769-2-2. PMC 4230743. PMID 25408874.] See also: Immunologic adjuvant
Peptidoglycan from Staphylococcus aureus was used as a vaccine to protect mice, showing that after vaccine injection for 40 weeks, the mice survived from S. aureus challenge at an increased lethal dose.
- Capparelli R, Nocerino N, Medaglia C, Blaiotta G, Bonelli P, Iannelli D (2011-12-01). Cardona PJ (ed.). “The Staphylococcus aureus peptidoglycan protects mice against the pathogen and eradicates experimentally induced infection”. PLOS ONE. 6 (12): e28377. Bibcode:2011PLoSO…628377C. doi:10.1371/journal.pone.0028377. PMC 3228750. PMID 22145040.
Inhibition and degradation
Some antibacterial drugs such as penicillin interfere with the production of peptidoglycan by binding to bacterial enzymes known as penicillin-binding proteins or DD-transpeptidases.
- Salton MR, Kim KS (1996). “Structure”. In Baron S, et al. (eds.). Structure. In: Baron’s Medical Microbiology (4th ed.). Univ of Texas Medical Branch. ISBN 978-0-9631172-1-2. PMID 21413343.
Penicillin-binding proteins form the bonds between oligopeptide crosslinks in peptidoglycan. For a bacterial cell to reproduce through binary fission, more than a million peptidoglycan subunits (NAM-NAG+oligopeptide) must be attached to existing subunits.
- Bauman R (2007). 2nd (ed.). Microbiology with Diseases by Taxonomy. Benjamin Cummings. ISBN 978-0-8053-7679-1.
Mutations in genes coding for transpeptidases that lead to reduced interactions with an antibiotic are a significant source of emerging antibiotic resistance.
- Spratt BG (April 1994). “Resistance to antibiotics mediated by target alterations”. Science. 264 (5157): 388–393. Bibcode:1994Sci…264..388S. doi:10.1126/science.8153626. PMID 8153626. S2CID 30578841.
Since peptidoglycan is also lacking in L-form bacteria and in mycoplasmas, both are resistant against penicillin.
Other steps of peptidoglycan synthesis can also be targeted. The topical antibiotic bacitracin targets the utilization of C55-isoprenyl pyrophosphate. Lantibiotics, which includes the food preservative nisin, attack lipid II.
- Sarkar P, Yarlagadda V, Ghosh C, Haldar J (March 2017). “A review on cell wall synthesis inhibitors with an emphasis on glycopeptide antibiotics”. MedChemComm. 8 (3): 516–533. doi:10.1039/c6md00585c. PMC 6072328. PMID 30108769.
Lysozyme, which is found in tears and constitutes part of the body’s innate immune system exerts its antibacterial effect by breaking the β-(1,4)-glycosidic bonds in peptidoglycan (see above). Lysozyme is more effective in acting against Gram-positive bacteria, in which the peptidoglycan cell wall is exposed, than against Gram-negative bacteria, which have an outer layer of LPS covering the peptidoglycan layer.
- Murphy K (2017). Janeway’s immunobiology. Casey Weaver, Charles Janeway (9th ed.). New York. pp. 45, 96–98. ISBN 978-0-8153-4505-3. OCLC 933586700.
Several bacterial peptidoglycan modifications can result in resistance to degradation by lysozyme. Susceptibility of bacteria to degradation is also considerably affected by exposure to antibiotics. Exposed bacteria synthesize peptidoglycan that contains shorter sugar chains that are poorly crosslinked and this peptidoglycan is then more easily degraded by lysozyme.
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
See also
References
- Woese CR, Kandler O, Wheelis ML (June 1990). “Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya”. Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–4579. Bibcode:1990PNAS…87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- Madigan, Michael T.; Martinko, John M.; Bender, Kelly S.; Buckley, Daniel H.; Stahl, David A. (2015). Brock Biology of Microorganisms (14 ed.). Boston: Pearson Education Limited. pp. 66–67. ISBN 978-1-292-01831-7.
- Mehta A (20 March 2011). “Animation of Synthesis of Peptidoglycan Layer”. PharmaXChange.info.
- Belgrave AM, Wolgemuth CW (June 2013). “Elasticity and biochemistry of growth relate replication rate to cell length and cross-link density in rod-shaped bacteria”. Biophysical Journal. 104 (12): 2607–2611. Bibcode:2013BpJ…104.2607B. doi:10.1016/j.bpj.2013.04.028. PMC 3686348. PMID 23790368.
- Purcell A (18 March 2016). “Bacteria”. Basic Biology.
- Hogan CM (12 October 2014). “Bacteria”. In Draggan S, Cleveland CJ (eds.). Encyclopedia of Earth. Washington DC: National Council for Science and the Environment.
- Salton MR, Kim KS (1996). “Structure”. In Baron S, et al. (eds.). Structure. In: Baron’s Medical Microbiology (4th ed.). Univ of Texas Medical Branch. ISBN 978-0-9631172-1-2. PMID 21413343.
- Demchick P, Koch AL (February 1996). “The permeability of the wall fabric of Escherichia coli and Bacillus subtilis”. Journal of Bacteriology. 178 (3): 768–773. doi:10.1128/jb.178.3.768-773.1996. PMC 177723. PMID 8550511.
- Coico R (October 2005). “Gram Staining”. In Coico R, Kowalik T, Quarles J, Stevenson B (eds.). Current Protocols in Microbiology. Vol. Appendix 3. Hoboken, NJ, USA: John Wiley & Sons, Inc. pp. mca03cs00. doi:10.1002/9780471729259.mca03cs00. ISBN 978-0-471-72925-9. PMID 18770544. S2CID 32452815.
- Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS (December 2008). “Cell shape and cell-wall organization in Gram-negative bacteria”. Proceedings of the National Academy of Sciences of the United States of America. 105 (49): 19282–19287. Bibcode:2008PNAS..10519282H. doi:10.1073/pnas.0805309105. PMC 2592989. PMID 19050072.
- Ryan KJ, Ray CG, eds. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 978-0-8385-8529-0.
- Schleifer KH, Kandler O (December 1972). “Peptidoglycan types of bacterial cell walls and their taxonomic implications”. Bacteriological Reviews. 36 (4): 407–477. doi:10.1128/MMBR.36.4.407-477.1972. PMC 408328. PMID 4568761.
- Kandler O, Hippe H (May 1977). “Lack of peptidoglycan in the cell walls of Methanosarcina barkeri”. Archives of Microbiology. 113 (1–2): 57–60. doi:10.1007/BF00428580. PMID 889387. S2CID 19145374.
- Kandler O, König H (April 1998). “Cell wall polymers in Archaea (Archaebacteria)”. Cellular and Molecular Life Sciences. 54 (4): 305–308. doi:10.1007/s000180050156. PMID 9614965. S2CID 13527169.
- Kandler G, Kandler O (1954). (Article in English available). “[Studies on morphology and multiplication of pleuropneumonia-like organisms and on bacterial L-phase, I. Light microscopy]” [Studies on morphology and multiplication of pleuropneumonia-like organisms and on bacterial L-phase, I. Light microscopy (now mycoplasmas and L-form bacteria)]. Archiv für Mikrobiologie (in German). 21 (2): 178–201. doi:10.1007/BF01816378. PMID 14350641. S2CID 21257985.
- Leaver M, Domínguez-Cuevas P, Coxhead JM, Daniel RA, Errington J (February 2009). [see also Erratum, 23 July 2009, Nature, vol. 460, p.538]. “Life without a wall or division machine in Bacillus subtilis”. Nature. 457 (7231): 849–853. Bibcode:2009Natur.457..849L. doi:10.1038/nature07742. PMID 19212404. S2CID 4413852.
- Kandler O (1994). “The early diversification of life”. In Bengtson S (ed.). Early Life on Earth. Nobel Symposium 84. New York: Columbia U.P. pp. 221–270. ISBN 9780231080880.
- Kandler O (1998). “The early diversification of life and the origin of the three domains: A proposal”. In Wiegel J, Adams MW (eds.). Thermophiles: The keys to molecular evolution and the origin of life?. London: Taylor and Francis Ltd. pp. 19–31. ISBN 978-0-203-48420-3.
- “The Prokaryotic Cell: Bacteria”. Archived from the original on 26 July 2010. Retrieved 1 May 2011.
- White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.
- Otten C, Brilli M, Vollmer W, Viollier PH, Salje J (January 2018). “Peptidoglycan in obligate intracellular bacteria”. Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880. PMC 5814848. PMID 29178391.
- Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N (July 2014). “Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis”. Science. 345 (6193): 220–222. Bibcode:2014Sci…345..220S. doi:10.1126/science.1254522. PMC 4163187. PMID 25013077.
- Madigan MT, Martinko JM, Dunlap PV, Clark DP (2009). Brock Biology of Microorganisms (12th ed.). San Francisco, CA: Pearson/Benjamin Cummings.
- König H, Kandler O, Hammes W (January 1989). “Biosynthesis of pseudomurein: isolation of putative precursors from Methanobacterium thermoautotrophicum”. Canadian Journal of Microbiology. 35 (1): 176–181. doi:10.1139/m89-027. PMID 2720492.
- Wolf AJ, Underhill DM (April 2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews. Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
- Bersch KL, DeMeester KE, Zagani R, Chen S, Wodzanowski KA, Liu S, et al. (April 2021). “Bacterial Peptidoglycan Fragments Differentially Regulate Innate Immune Signaling”. ACS Central Science. 7 (4): 688–696. doi:10.1021/acscentsci.1c00200. PMC 8155477. PMID 34056099.
- Bastos PA, Wheeler R, Boneca IG (January 2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
- Sun Q, Liu X, Li X (February 2022). “Peptidoglycan-based immunomodulation”. Applied Microbiology and Biotechnology. 106 (3): 981–993. doi:10.1007/s00253-022-11795-4. PMID 35076738. S2CID 246276803.
- Liang Y, Yang L, Wang Y, Tang T, Liu F, Zhang F (December 2022). “Peptidoglycan recognition protein SC (PGRP-SC) shapes gut microbiota richness, diversity and composition by modulating immunity in the house fly Musca domestica”. Insect Molecular Biology. 32 (2): 200–212. doi:10.1111/imb.12824. PMID 36522831. S2CID 254807823.
- Gonzalez-Santana A, Diaz Heijtz R (August 2020). “Bacterial Peptidoglycans from Microbiota in Neurodevelopment and Behavior” (PDF). Trends in Molecular Medicine. 26 (8): 729–743. doi:10.1016/j.molmed.2020.05.003. PMID 32507655. S2CID 219539658.
- Murphy K (2017). Janeway’s immunobiology. Casey Weaver, Charles Janeway (9th ed.). New York. pp. 45, 96–98. ISBN 978-0-8153-4505-3. OCLC 933586700.
- Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, Golenbock D (July 1999). “Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2”. Journal of Immunology. 163 (1): 1–5. doi:10.4049/jimmunol.163.1.1. PMID 10384090. S2CID 23630870.
- Capparelli R, Nocerino N, Medaglia C, Blaiotta G, Bonelli P, Iannelli D (2011-12-01). Cardona PJ (ed.). “The Staphylococcus aureus peptidoglycan protects mice against the pathogen and eradicates experimentally induced infection”. PLOS ONE. 6 (12): e28377. Bibcode:2011PLoSO…628377C. doi:10.1371/journal.pone.0028377. PMC 3228750. PMID 22145040.
- Bauman R (2007). 2nd (ed.). Microbiology with Diseases by Taxonomy. Benjamin Cummings. ISBN 978-0-8053-7679-1.
- Spratt BG (April 1994). “Resistance to antibiotics mediated by target alterations”. Science. 264 (5157): 388–393. Bibcode:1994Sci…264..388S. doi:10.1126/science.8153626. PMID 8153626. S2CID 30578841.
- Sarkar P, Yarlagadda V, Ghosh C, Haldar J (March 2017). “A review on cell wall synthesis inhibitors with an emphasis on glycopeptide antibiotics”. MedChemComm. 8 (3): 516–533. doi:10.1039/c6md00585c. PMC 6072328. PMID 30108769.
Further reading
- Bastos, Paulo A D; Wheeler, Richard; Boneca, Ivo G (2021). “Uptake, recognition and responses to peptidoglycan in the mammalian host”. FEMS Microbiology Reviews. 45 (1): fuaa044. doi:10.1093/femsre/fuaa044. PMC 7794044. PMID 32897324.
- Wolf, Andrea J.; Underhill, David M. (2018). “Peptidoglycan recognition by the innate immune system”. Nature Reviews Immunology. 18 (4): 243–254. doi:10.1038/nri.2017.136. PMID 29292393. S2CID 3894187.
- Gonzalez-Santana, Ayoze; Diaz Heijtz, Rochellys (2020). “Bacterial Peptidoglycans from Microbiota in Neurodevelopment and Behavior”. Trends in Molecular Medicine. 26 (8): 729–743. doi:10.1016/j.molmed.2020.05.003. PMID 32507655.
- Pohlschroder, Mechthild; Pfeiffer, Friedhelm; Schulze, Stefan; Halim, Mohd Farid Abdul (1 September 2018). “Archaeal cell surface biogenesis”. FEMS Microbiology Reviews. 42 (5): 694–717. doi:10.1093/femsre/fuy027. PMC 6098224. PMID 29912330.
Leave a Reply