DMRT1
Doublesex and mab-3 related transcription factor 1, also known as DMRT1, is a protein which in humans is encoded by the DMRT1 gene.
- “Entrez Gene: DMRT1 doublesex and mab-3 related transcription factor 1”.
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D (February 1998). “Evidence for evolutionary conservation of sex-determining genes”. Nature. 391 (6668): 691–5. Bibcode:1998Natur.391..691R. doi:10.1038/35618. PMID 9490411. S2CID 11414843.
- Raymond CS, Parker ED, Kettlewell JR, Brown LG, Page DC, Kusz K, Jaruzelska J, Reinberg Y, Flejter WL, Bardwell VJ, Hirsch B, Zarkower D (June 1999). “A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators”. Hum. Mol. Genet. 8 (6): 989–96. doi:10.1093/hmg/8.6.989. PMID 10332030.
Function
DMRT1 is a dose sensitive transcription factor protein that regulates Sertoli cells and germ cells. The DMRT1 gene is located at the end of the 9th chromosome. This gene is found in a cluster with two other members of the gene family, having in common a zinc finger-like DNA-binding motif (DM domain). The DM domain is an ancient, conserved component of the vertebrate sex-determining pathway that is also a key regulator of male development in flies and nematodes, and is found to be the key sex-determining factor in chickens. The majority of DMRT1 protein is located in the testicular cord and Sertoli cells, with a small amount in the germ cells.
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH (August 2009). “The avian Z-linked gene DMRT1 is required for male sex determination in the chicken”. Nature. 461 (7261): 267–71. Bibcode:2009Natur.461..267S. doi:10.1038/nature08298. PMID 19710650. S2CID 4413389.
Defective testicular development and XY feminization occur when this gene is hemizygous. Two copies of the DMRT1 gene are required for normal sexual development. When a DMRT1 gene is lost, the most common disease is chromosome 9p deletion, which causes abnormal testicular formation and feminization. The DMRT1 gene is critical for male sex determination; without this gene the female characteristic takes over and male characteristic is slight or non-existent.
When DMRT1 is knocked out in mice, the mice showed changes in both Sertoli and germ cells soon after formation of the gonadal ridge. The main defects associated with DMRT1 knockout were developmental arrest, excess proliferation of germ cells, and failure to undergo meiosis, mitosis, or migration. Thus, the knockout model shows that loss of the DMRT1 gene is associated with incomplete germ cell development leading to infertility, abnormal testicular formation, and/or feminization of the affected individual.[citation needed] Induced knockout of DMRT1 in adult male mice has been found to cause transdifferentiation of somatic cells in the testis to the equivalent cell types that would ordinarily be found in the ovary. Conversely, conditional expression of DMRT1 in the gonad of female mice caused the apparent transdifferentiation of ovarian somatic (GRANULOSA) cells to the equivalent cell type (SERTOLI) ordinarily found in males.
- Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D (July 2011). “DMRT1 prevents female reprogramming in the postnatal mammalian testis”. Nature. 476 (7358): 101–4. doi:10.1038/nature10239. PMC 3150961. PMID 21775990.
- Lindeman RE, Gearhart MD, Minkina A, Krentz AD, Bardwell VJ, Zarkower D (March 2015). “Sexual cell-fate reprogramming in the ovary by DMRT1”. Curr Biol. 25 (6): 764–71. doi:10.1016/j.cub.2015.01.034. PMC 4366330. PMID 25683803.
In molecular biology the DM domain is a protein domain first discovered in the DOUBLESEX proteins of Drosophila melanogaster and is also seen in C. elegans and mammalian proteins. In D. melanogaster the doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. These proteins are believed to function as transcription factors on downstream sex-determination genes, especially on NEUROBLAST differentiation and YOLK PROTEIN genes transcription.
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D (February 1998). “Evidence for evolutionary conservation of sex-determining genes”. Nature. 391 (6668): 691–5. Bibcode:1998Natur.391..691R. doi:10.1038/35618. PMID 9490411. S2CID 11414843.
- Erdman SE, Chen HJ, Burtis KC (December 1996). “Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins”. Genetics. 144 (4): 1639–52. doi:10.1093/genetics/144.4.1639. PMC 1207715. PMID 8978051.
- Burtis KC, Coschigano KT, Baker BS, Wensink PC (September 1991). “The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer”. EMBO J. 10 (9): 2577–82. doi:10.1002/j.1460-2075.1991.tb07798.x. PMC 452955. PMID 1907913.
- Shen MM, Hodgkin J (September 1988). “mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans”. Cell. 54 (7): 1019–31. doi:10.1016/0092-8674(88)90117-1. PMID 3046751. S2CID 1386352.
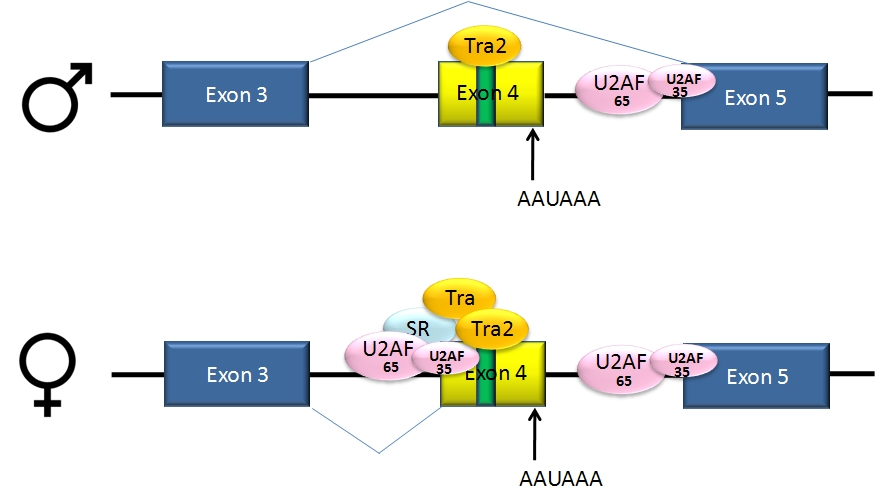
Doublesex (dsx) is a gene that is involved in the sex determination system of many insects including the fruit fly Drosophila melanogaster. The gene is expressed in both male and female flies and is subject to alternative splicing, producing the protein isoforms dsxf in females and the longer dsxm in males. The production of dsxf is caused by the presence of the female-specific version of the transformer (tra) gene. In a sense, the isoform of dsx informs a cell about the organism’s sex; for instance, female genitals only develop if dsxf is present. In conjunction with the gene fruitless, dsx also causes differences in the brain structure and behavior of males and females. Although the details of sex determination differ in the various species, there is a gene related to dsx in vertebrates (DMRT1) and in nematodes (MAB-3). All of these are transcription factors with a zinc finger DNA-binding domain (known as the DM domain) and are involved in sex-specific differentiation.
- Verhulst EC, van de Zande L, Beukeboom LW (August 2010). “Insect sex determination: it all evolves around transformer”. Current Opinion in Genetics & Development. 20 (4): 376–83. doi:10.1016/j.gde.2010.05.001. hdl:11370/2174764d-bd1b-4e1f-b142-90aced0c3e55. PMID 20570131. S2CID 205003182.
- Gilbert SF (2000), “Chromosomal Sex Determination in Drosophila“, Developmental Biology (6 ed.), Sinauer Associates
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF (April 2010). “Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster“. Nature Neuroscience. 13 (4): 458–66. doi:10.1038/nn.2515. PMC 3092424. PMID 20305646. — Lay summary: “‘Doublesex’ gene key to determining fruit fly gender”. ScienceDaily. 2022-02-03. Retrieved 2022-02-05.
The DM domain binds DNA as a dimer, allowing the recognition of pseudopalindromic sequences. The NMR analysis of the DSX DM domain revealed a novel zinc module containing ‘intertwined’ CCHC and HCCC zinc-binding sites. The recognition of the DNA requires the carboxy-terminal basic tail which contacts the minor groove of the target sequence.
- Yi W, Zarkower D (February 1999). “Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms”. Development. 126 (5): 873–81. doi:10.1242/dev.126.5.873. PMID 9927589.
- Zhu L, Wilken J, Phillips NB, Narendra U, Chan G, Stratton SM, Kent SB, Weiss MA (July 2000). “Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers”. Genes Dev. 14 (14): 1750–64. doi:10.1101/gad.14.14.1750. PMC 316782. PMID 10898790.
- Erdman SE, Chen HJ, Burtis KC (December 1996). “Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins”. Genetics. 144 (4): 1639–52. doi:10.1093/genetics/144.4.1639. PMC 1207715. PMID 8978051.
Proteins with this domain
Proteins with the DM domain are found in many model organisms. Many C. elegans Mab proteins contain this domain, the best-known one being mab-3. Human proteins containing this domain include DMRT1, DMRT2, DMRT3, DMRTA1, DMRTA2, DMRTB1, and DMRTC2; each of these have a mouse homolog.
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D (February 1998). “Evidence for evolutionary conservation of sex-determining genes”. Nature. 391 (6668): 691–5. Bibcode:1998Natur.391..691R. doi:10.1038/35618. PMID 9490411. S2CID 11414843.
- “Proteins matched: DM DNA-binding domain (IPR001275) filtered by species (Homo sapiens)”. InterPro.
DMRT1 homologs have an additional common domain C-terminal to the DM domain. This domain is only found in bony vertebrates, and neither its structure nor function is unknown. Jpred predicts the human version of the section to be mostly coils; it also suggests a weak similarity to PDB: 6BO4 by BLAST. (TRPV2 pore turret?)
- “Family: Dmrt1 (PF12374)”. Pfam.
- “Jpred results (MTECSGTSQPPPASVPTTAASEGRMVIQDIPAVTSRGHVENTPD)”. www.compbio.dundee.ac.uk. Archived from the original on 10 April 2019. Retrieved 10 April 2019.
DMRTA proteins have an additional motif in their C-termina. This motif, ubiquitous in eukaryotes, has an unknown function. It is similar in sequence to some UBIQUITIN-associated motifs.
- Species: DMRTA motif (IPR005173)”. InterPro. Retrieved 10 April 2019.
What brought us here
Mammals have two genes (SRY and DMT1) for testis formation-androgenesis, an anti-testis gene, DAX1, an anti-Müllerian duct hormone, and steroid sex hormones.
- De Loof, A, and R Huybrechts. “”Insects do not have sex hormones”: a myth?.” General and comparative endocrinology vol. 111,3 (1998): 245-60. doi:10.1006/gcen.1998.7101
So I’m a little confused. This one does seem to be more on target than DMT1…which I’m keeping because interesting and mentioned in at least one other note…but the chart at the bottom says (birds). I guess maybe dip into more than the abstract for details. 🙂
References
- GRCh38: Ensembl release 89: ENSG00000137090 – Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000024837 – Ensembl, May 2017
- “Human PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Mouse PubMed Reference:”. National Center for Biotechnology Information, U.S. National Library of Medicine.
- “Entrez Gene: DMRT1 doublesex and mab-3 related transcription factor 1”.
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D (February 1998). “Evidence for evolutionary conservation of sex-determining genes”. Nature. 391 (6668): 691–5. Bibcode:1998Natur.391..691R. doi:10.1038/35618. PMID 9490411. S2CID 11414843.
- Raymond CS, Parker ED, Kettlewell JR, Brown LG, Page DC, Kusz K, Jaruzelska J, Reinberg Y, Flejter WL, Bardwell VJ, Hirsch B, Zarkower D (June 1999). “A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators”. Hum. Mol. Genet. 8 (6): 989–96. doi:10.1093/hmg/8.6.989. PMID 10332030.
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH (August 2009). “The avian Z-linked gene DMRT1 is required for male sex determination in the chicken”. Nature. 461 (7261): 267–71. Bibcode:2009Natur.461..267S. doi:10.1038/nature08298. PMID 19710650. S2CID 4413389.
- Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D (July 2011). “DMRT1 prevents female reprogramming in the postnatal mammalian testis”. Nature. 476 (7358): 101–4. doi:10.1038/nature10239. PMC 3150961. PMID 21775990.
- Verhulst EC, van de Zande L, Beukeboom LW (August 2010). “Insect sex determination: it all evolves around transformer”. Current Opinion in Genetics & Development. 20 (4): 376–83. doi:10.1016/j.gde.2010.05.001. hdl:11370/2174764d-bd1b-4e1f-b142-90aced0c3e55. PMID 20570131. S2CID 205003182.
- Gilbert SF (2000), “Chromosomal Sex Determination in Drosophila“, Developmental Biology (6 ed.), Sinauer Associates
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF (April 2010). “Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster“. Nature Neuroscience. 13 (4): 458–66. doi:10.1038/nn.2515. PMC 3092424. PMID 20305646. — Lay summary: “‘Doublesex’ gene key to determining fruit fly gender”. ScienceDaily. 2022-02-03. Retrieved 2022-02-05.
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D (February 1998). “Evidence for evolutionary conservation of sex-determining genes”. Nature. 391 (6668): 691–5. Bibcode:1998Natur.391..691R. doi:10.1038/35618. PMID 9490411. S2CID 11414843.
- Erdman SE, Chen HJ, Burtis KC (December 1996). “Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins”. Genetics. 144 (4): 1639–52. doi:10.1093/genetics/144.4.1639. PMC 1207715. PMID 8978051.
- Burtis KC, Coschigano KT, Baker BS, Wensink PC (September 1991). “The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer”. EMBO J. 10 (9): 2577–82. doi:10.1002/j.1460-2075.1991.tb07798.x. PMC 452955. PMID 1907913.
- Shen MM, Hodgkin J (September 1988). “mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans”. Cell. 54 (7): 1019–31. doi:10.1016/0092-8674(88)90117-1. PMID 3046751. S2CID 1386352.
- Yi W, Zarkower D (February 1999). “Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms”. Development. 126 (5): 873–81. doi:10.1242/dev.126.5.873. PMID 9927589.
- Zhu L, Wilken J, Phillips NB, Narendra U, Chan G, Stratton SM, Kent SB, Weiss MA (July 2000). “Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers”. Genes Dev. 14 (14): 1750–64. doi:10.1101/gad.14.14.1750. PMC 316782. PMID 10898790.
- “Proteins matched: DM DNA-binding domain (IPR001275) filtered by species (Homo sapiens)”. InterPro.
- “Family: Dmrt1 (PF12374)”. Pfam.
- “Jpred results (MTECSGTSQPPPASVPTTAASEGRMVIQDIPAVTSRGHVENTPD)”. www.compbio.dundee.ac.uk. Archived from the original on 10 April 2019. Retrieved 10 April 2019.
- “Species: DMRTA motif (IPR005173)”. InterPro. Retrieved 10 April 2019.
- Lindeman RE, Gearhart MD, Minkina A, Krentz AD, Bardwell VJ, Zarkower D (March 2015). “Sexual cell-fate reprogramming in the ovary by DMRT1”. Curr Biol. 25 (6): 764–71. doi:10.1016/j.cub.2015.01.034. PMC 4366330. PMID 25683803.
- Anthony D. Krentza,b, Mark W. Murphya, Shinseog Kima,1, Matthew S. Cookc, Blanche Capelc, Rui Zhud, Angabin Matind, Aaron L. Sarvere, Keith L. Parkerf, Michael D. Griswoldg, Leendert H. J. Looijengah, Vivian J. Bardwella and David Zarkower. “The DM Domain Protein DMRT1 Is a Dose-sensitive Regulator of Fetal Germ Cell Proliferation and Pluripotency.” The DM Domain Protein DMRT1 Is a Dose-sensitive Regulator of Fetal Germ Cell Proliferation and Pluripotency. PNAS, 29 Oct. 2009. Web. 12 Mar. 2014.
- Christopher S. Raymond1, Emily D. Parker2, Jae R. Kettlewell1, Laura G. Brown3, David C. Page3, Kamila Kusz4, Jadwiga Jaruzelska4, Yuri Reinberg5, Wendy L. Flejter6, Vivian J. Bardwell1,2, Betsy Hirsch7 and David Zarkower1. “Human Molecular Genetics.” A Region of Human Chromosome 9p Required for Testis Development Contains Two Genes Related to Known Sexual Regulators. Oxford Journal, n.d. Web. 28 Feb. 2014.
- Craig A. Smith, Kelly N. Roeszler, Thomas Ohnesorg, David M. Cummins, Peter G. Farlie, Timothy J. Doran & Andrew H. Sinclair. “The Avian Z-linked Gene DMRT1 Is Required for Male Sex Determination in the Chicken.” Nature.com. Nature, 26 Aug. 2009. Web. 12 Mar. 2014.
- “DMRT1 Gene.” – GeneCards. Crown Human Genome Center, Department of Molecular Genetics, the Weizmann Institute of Science,http://genome.ucsc.edu/. 23 Oct. 2013. Web. 12 Mar. 2014.
- Ning Lei, Kaori I. Hornbaker, Daren A. Rice, Tatiana Karpova, Valentine A. Agbor, and Leslie L. Heckert. “Sex-specific Differences in Mouse DMRT1 Expression Are Both Cell Type- and Stage-dependent during Gonad Development.” Sex-specific Differences in Mouse DMRT1 Expression Are Both Cell Type- and Stage-dependent during Gonad Development. NIH Public Access, 13 June 2007. Web. 12 Mar. 2014.
Further reading
- Smith CA, McClive PJ, Western PS, et al. (2000). “Conservation of a sex-determining gene”. Nature. 402 (6762): 601–2. doi:10.1038/45127. PMID 10604464. S2CID 4401771.
- Calvari V, Bertini V, De Grandi A, et al. (2000). “A new submicroscopic deletion that refines the 9p region for sex reversal”. Genomics. 65 (3): 203–12. doi:10.1006/geno.2000.6160. PMID 10857744.
- Muroya K, Okuyama T, Goishi K, et al. (2000). “Sex-determining gene(s) on distal 9p: clinical and molecular studies in six cases”. J. Clin. Endocrinol. Metab. 85 (9): 3094–100. doi:10.1210/jcem.85.9.6771. PMID 10999792. S2CID 22145136.
- Raymond CS, Murphy MW, O’Sullivan MG, et al. (2000). “Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation”. Genes Dev. 14 (20): 2587–95. doi:10.1101/gad.834100. PMC 316999. PMID 11040213.
- Casci T (2001). “It’s a guy thing”. Nat. Rev. Genet. 1 (3): 169. doi:10.1038/35042034. PMID 11252745. S2CID 205010812.
- Harrington JJ, Sherf B, Rundlett S, et al. (2001). “Creation of genome-wide protein expression libraries using random activation of gene expression”. Nat. Biotechnol. 19 (5): 440–5. doi:10.1038/88107. PMID 11329013. S2CID 25064683.
- Boyer A, Dornan S, Daneau I, et al. (2004). “Conservation of the function of DMRT1 regulatory sequences in mammalian sex differentiation”. Genesis. 34 (4): 236–43. doi:10.1002/gene.10158. PMID 12434333. S2CID 7891350.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). “Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences”. Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS…9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Humphray SJ, Oliver K, Hunt AR, et al. (2004). “DNA sequence and analysis of human chromosome 9”. Nature. 429 (6990): 369–74. Bibcode:2004Natur.429..369H. doi:10.1038/nature02465. PMC 2734081. PMID 15164053.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). “The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)”. Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Cheng HH, Ying M, Tian YH, et al. (2006). “Transcriptional diversity of DMRT1 (dsx- and mab3-related transcription factor 1) in human testis”. Cell Res. 16 (4): 389–93. doi:10.1038/sj.cr.7310050. PMID 16617334.
- *1. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. (PubMed id 10332030)1, 2, 3, 9 Raymond C.S…. Zarkower D. (1999)
- 2. Evidence for evolutionary conservation of sex-determining genes. (PubMed id 9490411)1, 2, 3, 9 Raymond C.S….Zarkower D. (1998)
- 3. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. (PubMed id 20543847)1, 2, 4 Turnbull C….Rahman N. (2010)
- 4. Transcriptional diversity of DMRT1 (dsx- and mab3-related transcription factor 1) in human testis. (PubMed id 16617334)1, 2, 9 Cheng H.H….Zhou R.J. (2006)
- 5. A new submicroscopic deletion that refines the 9p region for sex reversal. (PubMed id 10857744)1, 2, 9 Calvari V…. Guioli S. (2000)
- 6. A second independent locus within DMRT1 is associated with testicular germ cell tumor susceptibility. (PubMed id 21551455)1, 4 Kanetsky P.A….Nathanson K.L. (2011)
- 7. Personalized smoking cessation: interactions between nicotine dose, dependence and quit-success genotype score. (PubMed id 20379614)1, 4 Rose J.E….Uhl G.R. (2010)
- 8. DNA sequence and analysis of human chromosome 9. (PubMed id 15164053)1, 2 Humphray S.J…. Dunham I. (2004)
- 9. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC). (PubMed id 15489334)1,2 Gerhard D.S….Malek J. (2004)
- 10. The DM domain protein DMRT1 is a dose-sensitive regul ator of fetal germ cell proliferation and pluripotency. (PubMed id 20007774)1, 9 Krentz A.D….Zarkower D. (2009)
External links
- DMRT1+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- https://web.archive.org/web/20110720091853/http://www.dmrt1.umn.edu/
- Overview of all the structural information available in the PDB for UniProt: Q9Y5R6 (Doublesex- and mab-3-related transcription factor 1) at the PDBe-KB.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
| Transcription factors and intracellular receptors | |
|---|---|
| show(1) Basic domains | |
| show(2) Zinc finger DNA-binding domains | |
| show(3) Helix-turn-helix domains | |
| show(4) β-Scaffold factors with minor groove contacts | |
| show(0) Other transcription factors | |
| see also transcription factor/coregulator deficiencies |
| Sex determination and differentiation | |
|---|---|
| Overview | Sexual differentiation humans Development of the reproductive system gonads Mesonephric duct Paramesonephric duct |
| Genetic basis | Sex-determination system XY X0 ZW Temperature-dependent Haplodiploidy Sex chromosome X chromosome Y chromosome Sex determining gene: SRY (mammal) DMRT1 (birds) |
| See also | Hermaphrodite Intersex Disorders of sex development Sex reversal |


Leave a Reply