Structure of DNA repair protein XRCC4 aka X-ray repair cross-complementing protein 4
XRCC4 protein is a TETRAMER that resembles the shape of a DUMBBELL containing two globular ends separated by a long, thin stalk. The tetramer is composed of two dimers, and each dimer is made up of two similar subunits. The first subunit (L) contains amino acid residues 1 – 203 and has a longer
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is active in both non-dividing and proliferating cells.
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. It is called “non-homologous” because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair (HDR), which requires a&nb
Ku is a dimeric protein complex that binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair
Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer (two copies of the same protein bound to each other).[2] Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human
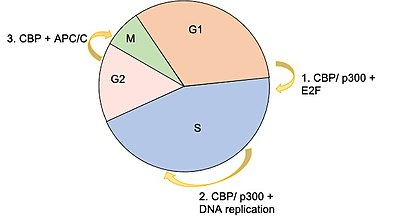
CBP has two critical mechanisms by which it is able to regulate gene expression: as an acetyltransferase, and as a protein scaffold
This gene is ubiquitously expressed and is involved in the transcriptional coactivation of many different transcription factors. CBP has two critical mechanisms by which it is able to regulate gene expression: as an acetyltransferase, and as a protein scaffold that helps recruit and construct the
Proteins shown to interact specifically with CBP (list)
ActrNuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300.Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG (April 2007). “Roles of CREB-binding protein (CBP)/p300 in respiratory epithelium tu
Surfeit human gene cluster
Surfeit is a human gene cluster that consists of a group of very tightly linked genes on chromosome 9 that do not share sequence similarity. Genes in this cluster are numbered 1 through 6: SURF1, SURF2, SURF3, SURF4, SURF5, and SURF6. Surfeit locus protein 1 (SURF1) Surfeit locus protein
Tryptophan tryptophylquinone (TTQ) formation
Tryptophan tryptophylquinone (TTQ) is an enzyme cofactor, generated by posttranslational modification of amino acids within the protein. Methylamine dehydrogenase (MADH), an amine dehydrogenase, requires TTQ for its catalytic function. From Wikipedia wher
Structural motif
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. Johansson, M.U. (23 July 2012). “Defining and searching for structural motif